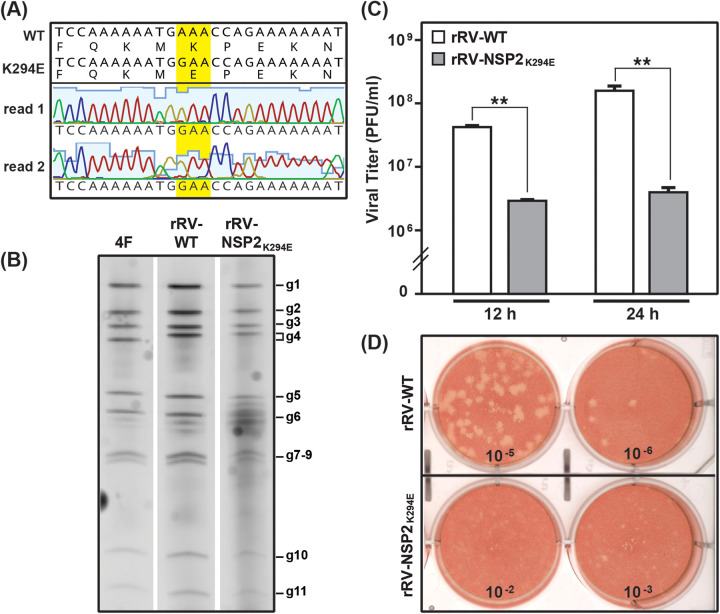
FIG 3.
Reverse genetics rescue and replication of rRV-NSP2K294E. (A) Sequence analysis of the rRV-NSP2K294E working stock. The NSP2-coding gene of rRV-NSP2K294E was amplified by RT-PCR and subjected to Sanger sequencing. The chromatograms of two independent sequencing reactions (read 1 and read 2) are shown across the engineered lesion only. Nucleotide and amino acid sequences of the unmodified wild-type (WT) pT7-NSP2-SA11 plasmid are shown above the consensus mutant virus sequence. The engineered codon is highlighted with yellow. (B) Electropherotyping. Viral dsRNA was extracted from laboratory strain SA11-4F (4F), rRV-WT, or rRV-NSP2K294E working viral stock and visualized following SDS-PAGE and SYBER gold staining. The positions of the 11 viral genome segments (g1 to g11) are labeled to the right of the gel. (C) Single-cycle replication assay. MA104 cells were infected at an MOI of 3 PFU per cell with either rRV-WT (white) or rRV-NSP2K294E (gray). Viral titers were determined by plaque assay. Error bars represent the standard error from the mean. Statistical significance was determined using a two-tailed, unpaired t test. Asterisks (**) indicate a P value of <0.001. (D) Images of representative plaque assay wells for rRV-WT (top) or rRV-NSP2K294E (bottom) at 9 days p.i. following neutral red overlay.