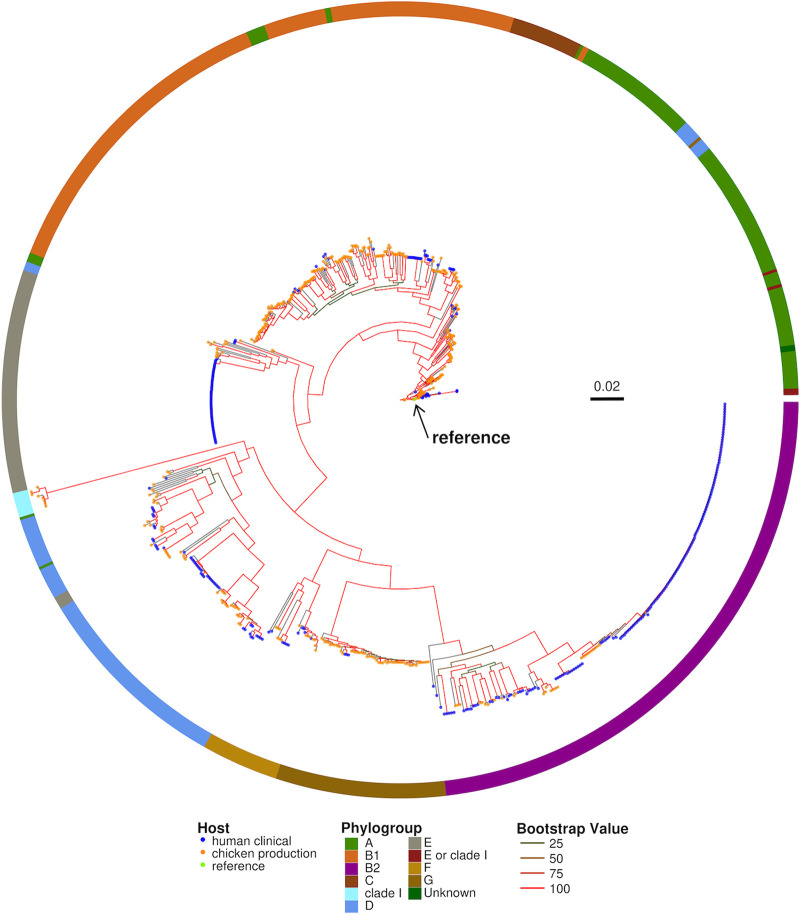
FIG 3.
Whole-genome SNP-based maximum-likelihood phylogenetic tree. SNPs found between the E. coli K12-MG1655 (accession U00096) reference genome, and each of the 791 E. coli isolates was used for alignment using Snippy v4.6.0 (Snippy 2018). The resulting whole-genome SNP alignment was used to construct maximum-likelihood phylogenetic trees under a GTR+GAMMA model using RAxML with 100 bootstraps (RAxMLHPC-PTHREADS, v8.2.12). The strength of nodal support is indicated by the branch color: red (100), orange (50> to ≤ 75), dark orange (25> to ≤ 50), and green (0> to ≤ 25). (Ring) Phylogroups identified using ClermonTyping v1.4.0. Phylogroups are labeled by color: A (green), B1 (orange), B2 (magenta), C (brown), clade I (aquamarine), D (blue), E (gray), E or clade I (maroon), F (gold), G (dark gold), and unknown (dark green).