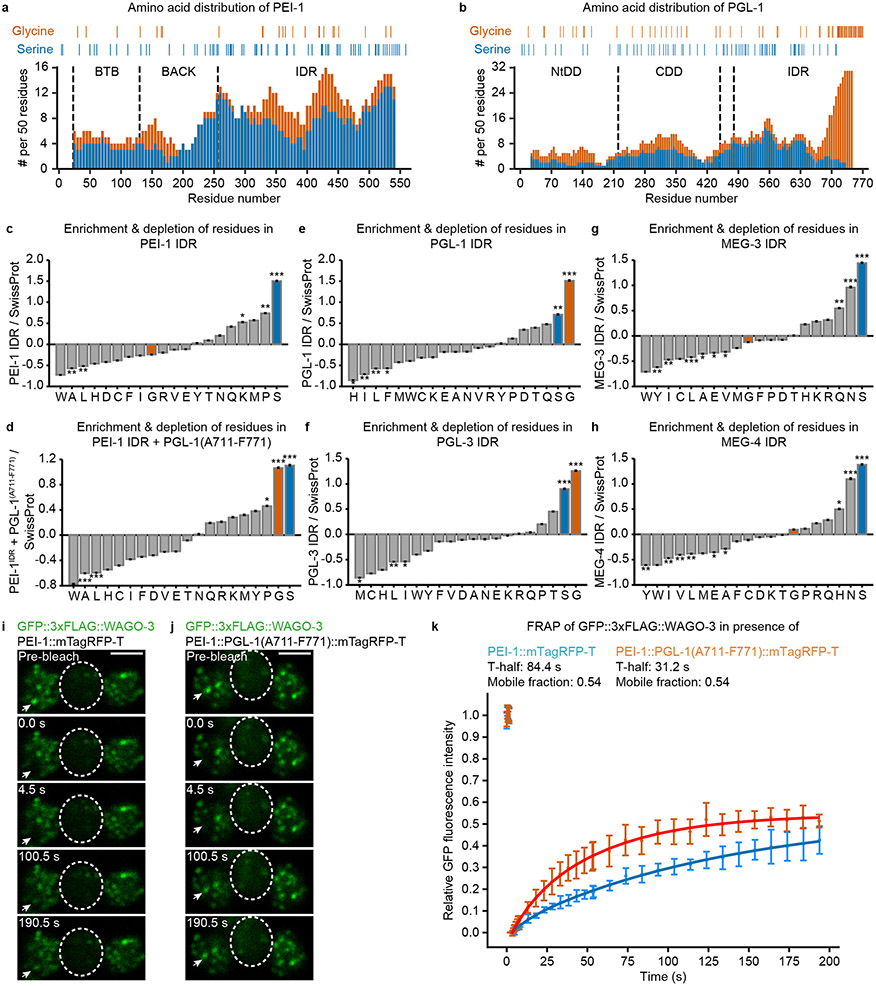
Extended Data Fig. 7. The amino acid composition of the PEI-1 IDR affects exchange dynamics of WAGO-3.
a-b, Occurrence of glycine and serine residues in PEI-1 (a) and PGL-1 (b) was counted in amino acid 50-mers, starting at position one, shifting 5 residues at a time, and displayed as stacked columns. Indicated residue positions in the diagrams are the mid-point of the 50-mer. Y-axes display number of relevant residues in amino acid 50-mers. X-axes indicate the position along the respective proteins. The various domains are indicated by vertical, dashed lines. NtDD and CDD indicate the N-terminal and C-terminal dimerization domains of PGL-1, respectively. The exact positions of glycine and serine residues for each protein are indicated above the stacked bar diagrams. IDR – intrinsically disordered region. c-h, Amino acid composition profiles of the intrinsically disordered region of the indicated proteins. Bars representing serine and glycine residues are highlighted in blue and orange, respectively. Panel d reflects a fusion between the PEI-1 IDR and the very C-terminal end of PGL-1(A711-F771). The profiles were generated using Composition profiler. Sequences were analyzed against the SwissProt database using 10,000 bootstrap iterations. Statistical significance was tested using the two sample t test (***: p ≤ 0.001, **: p ≤ 0.01, *: p ≤ 0.05, ns: p > 0.05). The exact P values are provided as source data. i-j, Time lapse images showing fluorescence recovery after photobleaching (FRAP) of GFP::3xFLAG::WAGO-3 localizing to PEI granules via PEI-1::mTagRFP-T (i) or via PEI-1::PGL-1(A711-F771)::mTagRFP-T (j) in isolated, male-derived budding spermatids. Residual bodies are marked by a dashed circle. Images represent two biologically independent experiments. Scale bars: 4 μm. k, FRAP recovery curves of GFP::3xFLAG::WAGO-3 localizing to PEI granules, containing indicated PEI-1 proteins, in male-derived budding spermatids. Normalized data is presented as mean +/− SD and was fitted to a double exponential curve (n = 5 granules pooled from one independent experiment). Source data are provided.