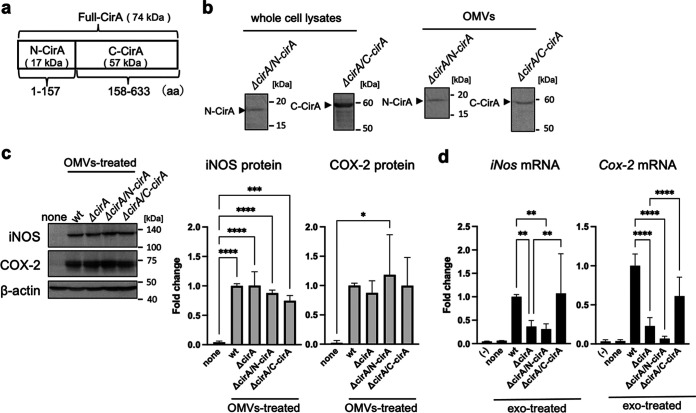
FIG 6.
C-terminal CirA caused exosome-mediated inflammatory responses. (a) The molecular weight and structure of N-terminal and C-terminal CirA protein (N-CirA and C-CirA) are shown. (b to d) OMVs were isolated from the culture medium of E. coli wild type (WT), cirA gene-deleted mutant strains (ΔcirA), and ΔcirA transformed with pTV-N-cirA vector or pTV-C-cirA vector (ΔcirA/N-cirA or ΔcirA/C-cirA) by ultracentrifugation. (b) The protein expression of N-CirA and C-CirA in whole-cell lysates and OMVs was measured by Western blotting. (c) Mouse RAW264.7 macrophages (Mφ) were incubated with OMVs (10 μg/7 mL) for 9 h. After the culture medium was replaced with fresh medium containing antibiotics, cells were incubated for 1 h. After replacement with fresh media containing antibiotics again, cells were incubated for 9 h. The protein expression of inducible nitric oxide synthase (iNOS) and cyclooxygenase-2 (COX-2) in whole-cell lysates was measured by Western blotting. β-Actin was used as loading control. CS Analyzer 3.0 was used for image analysis software. Relative intensity was expressed as the mean ± SD (n = 3) of at least three independent biological replicates. (d) Exosomes were isolated from cell culture medium by ultracentrifugation. RAW264.7 cells (recipient cells; 5 × 105 cells) were incubated with each exosome from donor cells (5 × 105 cells). After incubation for 12 h, the mRNA levels of iNos and Cox-2 were analyzed by quantitative real-time PCR and normalized to 18S rRNA. Values are expressed as the mean ± SD (n = 6 to 9) of at least three independent biological replicates. One-way ANOVA and Tukey’s test were used for statistical analysis; *, P < 0.05; **, P < 0.01; ***, P < 0.005; ****, P < 0.001.