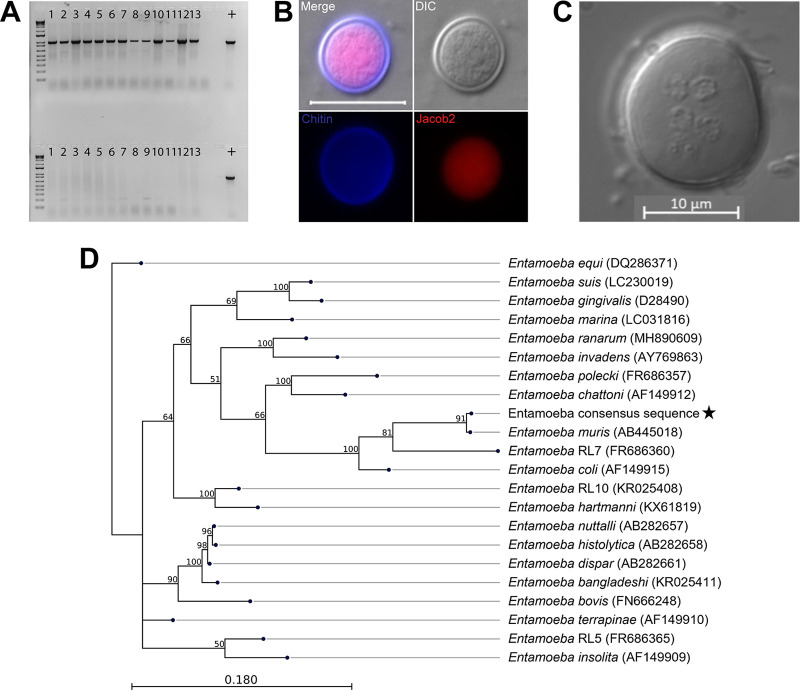
FIG 1.
Identification of a murine Entamoeba species. (A) Representative Pan-Entamoeba PCR for C57BL/6 mice (top) and Swiss Webster mice (bottom) within our vivarium. The loading control was murine GAPDH (data not shown). Each lane number represents a mouse cage. Positive control (+) is isolated genomic DNA from axenic Entamoeba histolytica culture. (B) Immunofluorescence assay staining for chitin (Calcofluor White), Jacob2 (1A4 antibody), scale bar represents 20 μm. (C) Representative phenotypic characterization of the number of nuclei of E. muris cysts (>4 nuclei). Image was taken using DIC. Scale bar represents 10 μm. (D) 18S phylogeny is based on a 1,033 bp alignment, including 27 published Entamoeba sequences, labeled as species name (NCBI accession), plus the Entamoeba found in our vivarium, indicated by a star. Numbers on branches are bootstrap values (%) based on 1,000 replicates (values >50% are shown). The scale bar indicates nucleotide substitutions per site.