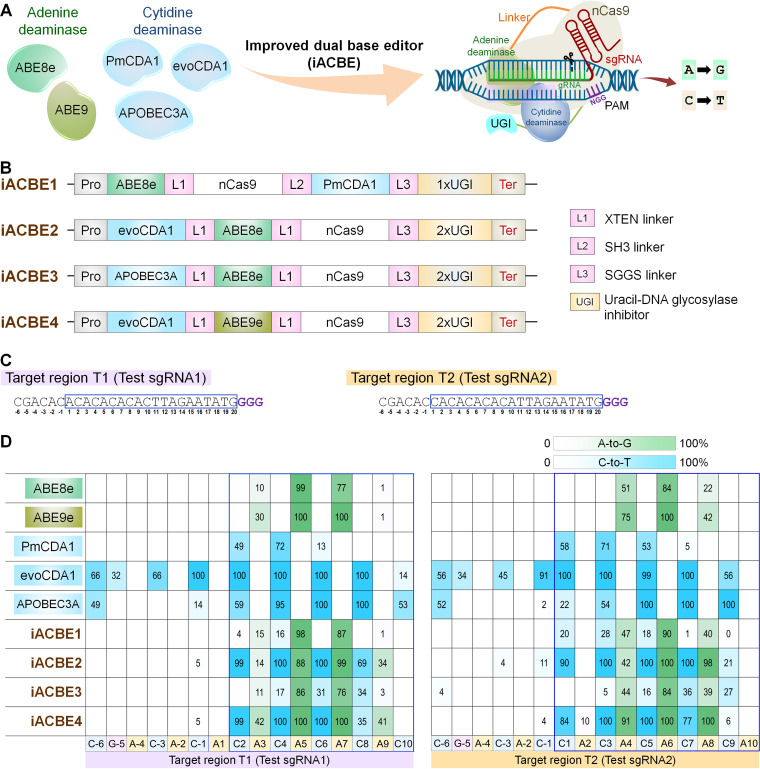
FIG 1.
Improved dual base editor (iACBE) systems induce synchronized A-to-G and C-to-T mutations in E. coli cells. (A) Deaminases used for generation of improved dual base editors (iACBEs). (B) Schematic representation of the plasmid vectors iACBEs tested in the work for iACBE development. Pro, pGlpT promoter; Ter, L3S2P21 terminator; nCas9, SpCas9 nickase with D10A mutation. Different deaminases are explained in the main text. L1, XTEN liner; L2, SH3 linker; L3, SGGS linker. (C) Two independent sgRNAs, including alternate Cs at even (test sgRNA1) and odd (test gRNA2) positions spanning from 1 to 10 in the 20 bp gRNA-spacer were expressed using pAtU6 promoter. The numbers are assigned by counting the distal base as 1 from the protospacer adjacent motif (PAM), i.e., GGG counting as 21 to 23. (D) Base editing (C-to-T and A-to-G) activities by single and dual base editors (iACBEs). Heat map values show the mean percentage of base conversion at different positions in target sites calculated from four independent biological replicates. Nucleotide bases located withing the 20 bp gRNA highlighted inside the blue box. The base conversion rate was estimated using the online EditR tool.