FIG 4.
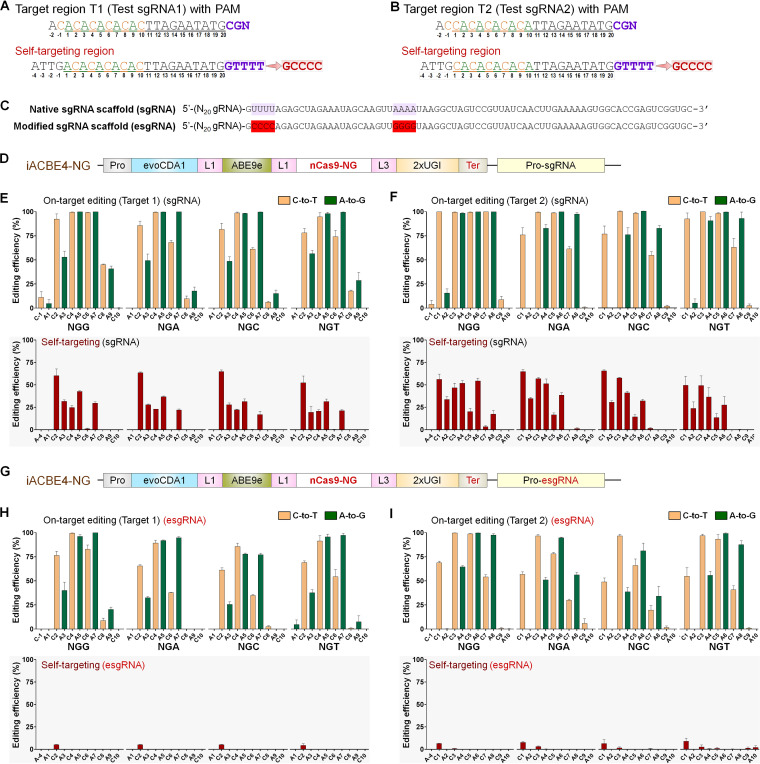
PAM-flexible SpCas9-NG nickase-based iACBE4-NG development for bacterial dual base editing. (A) The on-target site with protospacer adjacent motif (PAM) and self-targeting region in single guide RNA (sgRNA) cassette for Test sgRNA1 are shown. Editable As and Cs are highlighted in green and orange, respectively. The numbers are assigned by counting the distal base as 1 from the PAM, i.e., NGN counting as 21 to 23, where N is A/G/C/T. In the self-editing region, the native sgRNA scaffold starts with GTTTT (violet) and modified (esgRNA) starts with GCCCC (red). (B) The on-target and self-targeting regions of Test sgRNA2. (C) Nucleotide sequences of native and evolved sgRNAs. Nucleotides in native sgRNA colored in violet were changed in esgRNA (shaded in red). (D) Architecture of the plasmid construct illustrating iACBE4-NG and native sgRNA for testing editing features at NGN PAM motifs. L1, XTEN liner; L3, SGGS linker. (E and F) Concurrent A/C base editing activities by iACBE4-NG with native sgRNA cassettes at on-target and self-targeting regions. (G). Architecture of iACBE4-NG plasmid construct containing esgRNA. (H and I) On-target and self-target A/C base editing activities by iACBE4-NG consisting of esgRNA cassettes.