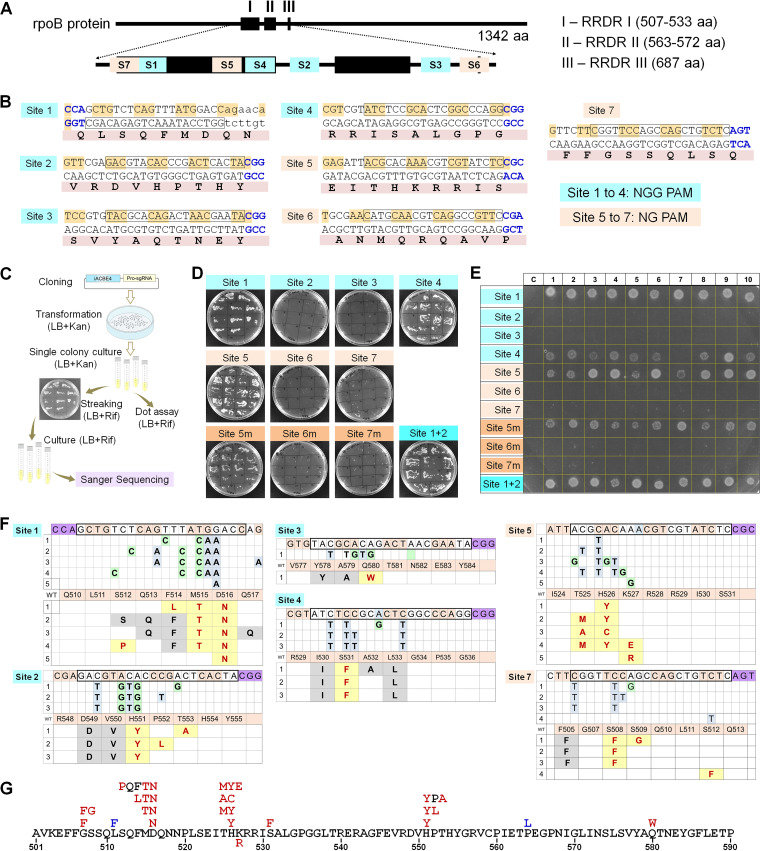
FIG 6.
Targeted mutagenesis of the rpoB gene by iACBE4 and iACBE4-NG identifies novel (previously unknown) mutations bestowing rifampicin resistance in E. coli. (A) Outline of rifampicin resistance-determining regions (RRDRs) within rpoB protein (black boxes) and distribution of intended target sites (S1 to S7) for targeted mutagenesis. (B) The DNA and corresponding amino acid sequences of target sites. Sites 1 to 4 contained protospacers (highlighted in box) with NGG as PAM (blue) and were targeted by iACBE4. Protospacers at sites 5 to 7 contained NGN as PAM motifs. (C) Experimental setup for generating rpoB mutants and screening for rifampicin resistance (RifR). (D and E) Screening of rpoB mutants performed by streak (D) and dot (E) assay on LB-agar plates with rifampicin. Cultures of individual clones streaked or dotted on plates were derived from the targeted mutagenesis by iACBE4 with native sgRNA scaffolds at sites 1 to 4. For sites 5 to 7, iACBE4-NG comprised of native scaffold (labeled as site 5, site 6, site 7) and modified (esgRNA) scaffold (labeled as site 5m, site 6m, site 7m). (F) Rifampicin-resistant rpoB alleles generated at different target sites. Multiple alleles with the same genotype are depicted only once, and nucleotide changes showing 20% or above are only considered in the analysis. WT indicates wild type amino acid sequence as a reference. Modified amino acids are highlighted in bold red with a yellow shade. Silent mutations showing editing at the DNA level but without altering amino acids are highlighted in a gray shade. (G) Distribution of mutations across 501 to 590 amino acid region of rpoB protein. Mutations found in the canonical BE window of iACBE are shown in red. Mutations found in the noncanonical iACBE window are shown in blue.