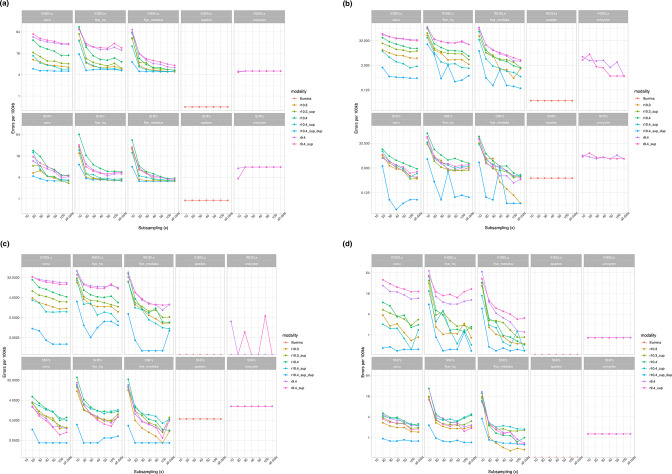
Fig. 7.
Impact of subsampling of long-read datasets on assembly accuracy. Presented here by species for Indels (top panels), and SNPs (lower panels). For ease of representation, only data for Flye assemblies polished with one round of Medaka are shown, as the effects of additional polishing was shown to be marginal for most modalities (Fig. S6, Table S7). Data for 10× long-read coverage is omitted for Canu assemblies as this coverage was considered too low for default settings and was unlikely to improve results. (a) E. coli (b) K. pneumoniae (chromosome only) (c) P. aeruginosa (d) S. aureus .