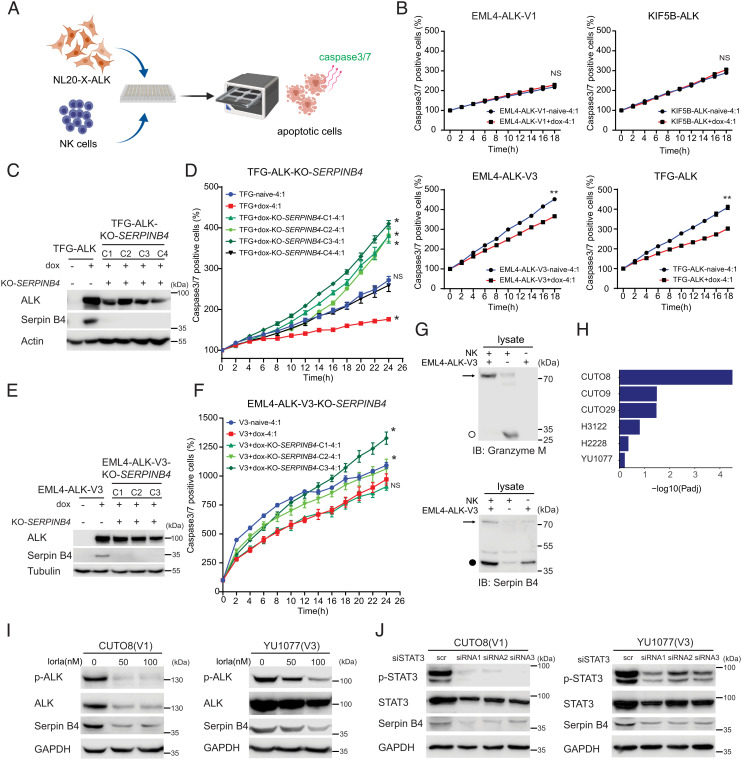
Fig. 7.
Serpin B4 expression rescues ALK expressing NL20 cells from NK cell-mediated cytotoxicity. (A) Schematic outline of the NK cell and NL20-ALK cell co-culture assay employed. NL20-ALK cells were primed with doxycycline for 2 d, prior to incubation with NK cells. Incucyte® Caspase-3/7 Dye was used as an indicator for cells undergoing apoptosis. Apoptotic signals (green fluorescence) were analyzed by an IncuCyte® Live Cell Analysis system. (B) NL20-ALK cells primed with doxycycline for 2 d were incubated with NK cells at an effector to target cell ratio (E:T) of 4:1. Caspase-3/7 positive cell percentages were calculated and normalized to 0 h using an IncuCyte® Live Cell Analysis system. Each data point represents mean ± SEM of normalized percentage conducted in triplicate (NS, non-significant; **P < 0.01; two-tailed paired t test). One typical experiment of three independent experiments is shown. (C) Immunoblotting analysis of Serpin B4 protein expression in parental NL20-TFG-ALK and four independent SERPINB4 CRISPR/CRISPR associated protein 9 (Cas9)-engineered KO clones (C1 to C4). Cell lysates were immunoblotted with ALK, Serpin B4, and Actin antibodies as indicated. (D) SERPINB4 KO NL20-TFG-ALK (C1 to C4) and parental control cells were primed with doxycycline (dox) for 2 d prior to co-culturing with NK cells and an E:T ratio of 4:1 and the percentage of caspase-3/7 positive cells calculated. Each data point represents mean ± SEM of normalized percentage conducted in triplicate (NS, non-significant; *P < 0.01; two-tailed paired t test). One typical experiment of three independent experiments is shown. (E) Immunoblotting analysis of Serpin B4 protein expression in parental NL20-EML4-ALK-V3 and three independent SERPINB4 CRISPR/Cas9-engineered KO clones (C1 to C3). Cell lysates were immunoblotted with ALK, Serpin B4, and Tubulin antibodies as indicated. (F) SERPINB4 KO NL20-EML4-ALK-V3 (C1 to C3) and parental control cells were primed with doxycycline (dox) for 2 d prior to co-culturing with NK cells and an E:T ratio of 4:1 and the percentage of caspase-3/7 positive cells calculated. Each data point represents mean ± SEM of normalized percentage conducted in triplicate (NS, non-significant; *P < 0.01; two-tailed paired t test). One typical experiment of three independent experiments is shown. (G) Immunoblotting analysis of co-incubated NK-92 and NL20-EML4-ALK-V3 lysates using Granzyme M and Serpin B4 antibodies indicates expression of Granzyme M (○, 27.5 kDa) in NK-92 cell lysates and Serpin B4 (●, 45 kDa) in NL20-EML4-ALK-V3 cell lysates. The formation of a Serpin B4 and Granzyme M complex (72.5 kDa) in the co-incubated lysates is indicated with an arrow. (H) Gene set enrichment for 27 inflammatory response genes (35) in DE genes in patient-derived cell lines treated with lorlatinib as indicated. GSEA was performed using Fisher’s exact test. Bars represent Padj values (–log10 scale). (I) CUTO8 and YU1077 were treated with lorlatinib (50 or 100 nM), and lysates were immunoblotted with pY1278-ALK, pan-ALK, Serpin B4, and GAPDH as indicated. Phospho-ALK was employed as readout of ALK inhibition. (J) CUTO8 and YU1077 cells were transfected with either control siRNA (scr) or siRNAs (1 to 3) targeting STAT3. Cell lysates were immunoblotted with pY705-STAT3, pan-STAT3, Serpin B4, and GAPDH antibodies as indicated.