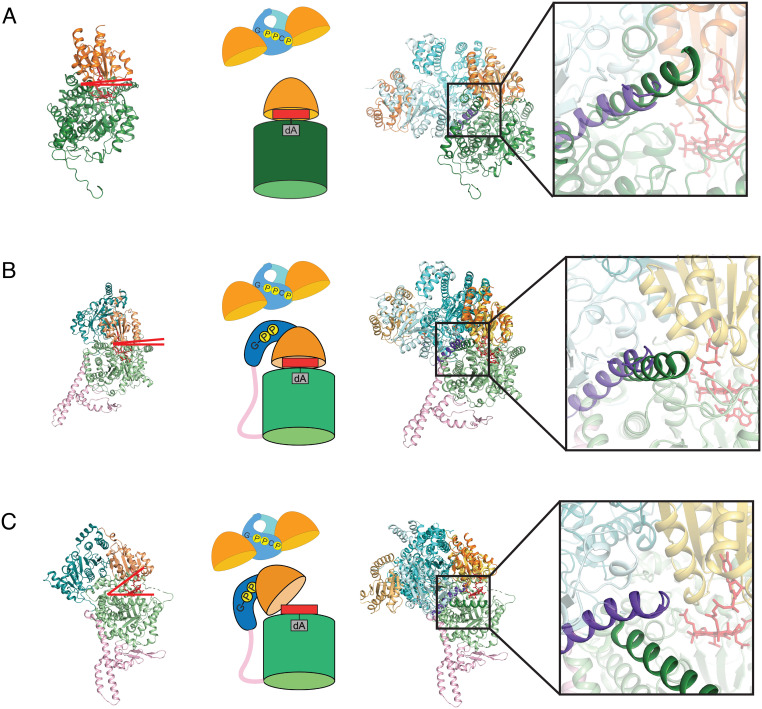
Fig. 6.
Superimposition of MeaB:MeMCMcbl:GMPPCP onto full-length mutase structures. (A) (Left) Ribbon drawing of PsMCM (PDB 2REQ) with substrate-binding domain (dark green) and Cbl-binding domain (light orange). Red lines indicate that there is no gap between domains; the Cbl is positioned into the barrel for catalysis. (Middle) Cartoon of MeaB:MeMCMcbl and an active protomer of PsMCM. (Right) Overlay of MeaB:MeMCMcbl with the PsMCM structure with Inset showing clash between an alpha helix of MeaB (residues 206 to 228 in purple) and the substrate binding domain (residues 441 to 459 in dark green) of PsMCM. (B) (Left) IcmF (PDB 4XC6) with substrate-binding domain (light green), Cbl-binding domain (light orange), G-protein domain (teal), and linker (pink). Red lines indicate that there is no gap between domains; the Cbl is positioned into the barrel for catalysis. (Middle) Cartoon of MeaB:MeMCMcbl and an active IcmF protomer. (Right) Overlay of MeaB:MeMCMcbl with the closed IcmF structure with Inset showing a clash between an alpha helix of MeaB (residues 206 to 228 in purple) and the substrate binding domain (residues 975 to 995 in dark green) of IcmF. (C) (Left) Ribbon drawing of open IcmF (PDB 4XC6) with colors indicated in (B). Red lines indicate a gap between domains is available for Cbl insertion. (Middle) Cartoon of MeaB:MeMCMcbl and an inactive protomer of IcmF. (Right) Overlay of MeaB:MeMCMcbl with the open protomer of IcmF with Inset showing that the helix of the substrate binding domain of IcmF (dark green) does not clash with the helix of MeaB (purple) when the mutase is “open”.