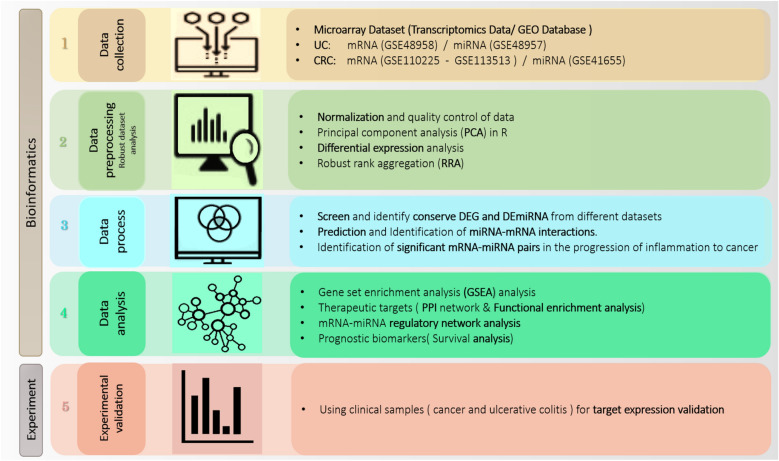
Figure 1.
Five sets of microarray data were downloaded and analyzed for differential identification. mRNAs, miRNAs are expressed in UC, CRC and normal tissues. By performing normalization and quality control, differential expression analysis of the data was performed and then the data was entered into RRA to find. The most significant DEGs and DE miRNAs were screened in all datasets. By predicting and identifying miRNA-mRNA interactions, important mRNA-miRNA pairs were identified in the progression of inflammation to cancer. To determine the key modules shared between CRC and UC, protein–protein interaction (PPI) network analyzes were performed and functional enrichment analysis was investigated. In addition, criteria and selection of common DEGs, conserved hub genes in the progression of inflammation to cancer were also obtained. Using DEmiRNAs, DEGs as well as conserved hub genes, the mRNA-miRNA regulatory network was constructed to find potential therapeutic targets. Finally, a comprehensive downstream analysis was performed to identify potential prognostic and diagnostic biomarkers through different approaches.