Figure 1.
Focused CRISPR-Cas9 kinome screen identifies Mvk and Pmvk dependencies in Bap1-deficient mesothelioma cells
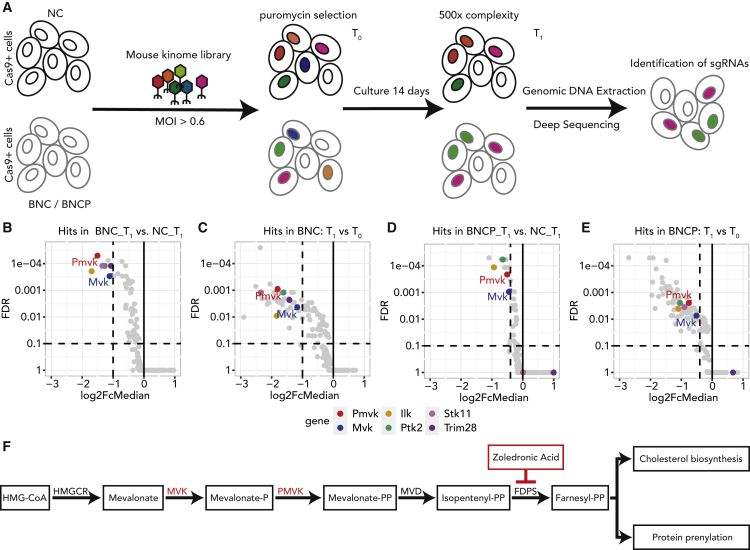
(A) Schematic representation of experimental workflow of the dropout kinome CRISPR-Cas9 screen in Bap1-deficient (BNC/BNCP) and proficient (NC) mesothelioma cell lines.
(B–E) Volcano plots showing the significantly dropped-out genes, (B) comparing BNC and NC at T1 (FDR ≤ 0.1, log2 fold change [FC] ≤ −1); (C) comparing T1 with T0 for BNC cells (FDR ≤ 0.1, log2FC ≤ −1); (D) comparing BNCP and NC at T1 (FDR ≤ 0.1, log2FC ≤ −0.4); and (E) comparing T1 with T0 for BNCP cells (FDR ≤ 0.1, log2FC ≤ −0.4). The top 6 hits from the screen are highlighted in color.
(F) Simplified schematic representation of the mevalonate pathway where primary hits from our screen are highlighted in red color. The inhibitor of FDPS, zoledronic acid, is highlighted as well.
See also Figure S1.