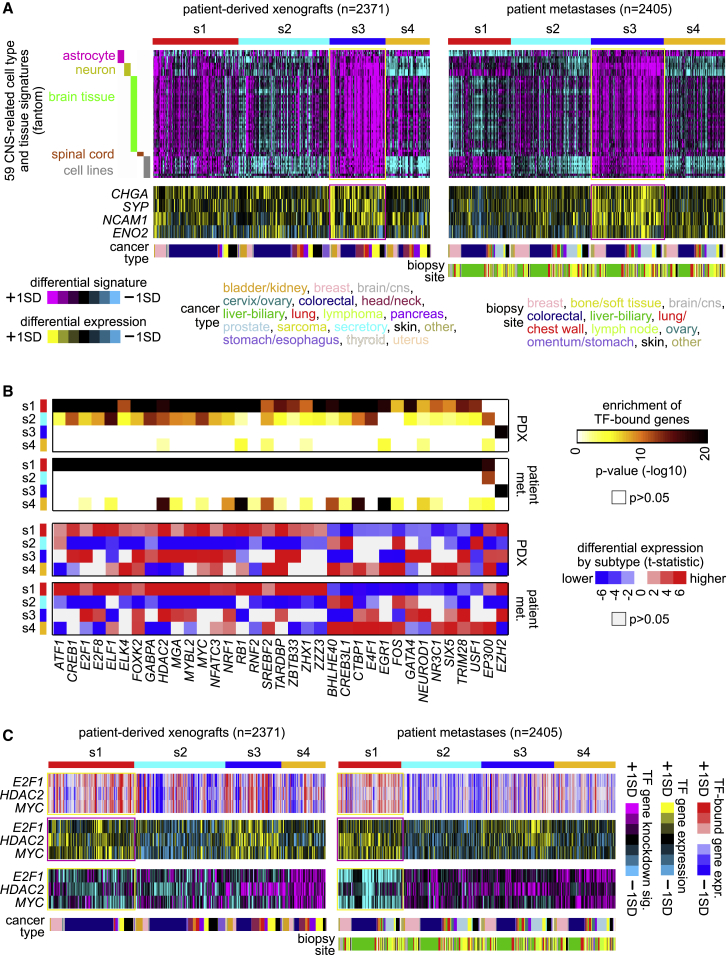
Figure 4.
Central nervous system (CNS) and transcription factor (TF) associations by subtype
(A) CNS associations by subtype. Heatmaps showing intersample correlations (purple, positive; cyan, negative) between mRNA profiles of tumors in PDX and patient metastasis compendium datasets (columns) and mRNA profiles of Fantom20 cell types or tissues related to the CNS (rows).
(B) Top TF associations by subtype. Encode21 data on TF binding was extracted for 158 TFs, with gene associations defined by binding within 2 kb upstream of the gene start. For the set of 35 TFs represented, there was both significant overlap (p < 1E−6, one-sided Fisher’s exact test or chi-square test) between the TF-bound genes and the genes overexpressed in the expression subtype and significantly higher or lower levels of the TF gene in that same subtype (p < 0.05 by t test), for both PDX and patient metastasis compendium datasets.
(C) Of the 35 TFs represented in (B), 10 were represented in an expression profiling dataset of siRNA knockdown of specific genes.22 For three TFs—E2F1, HDAC2, and MYC—the genes were highly expressed in the associated s1 subtype, with the corresponding siRNA knockdown signature scoring significantly negative (p < 1E−6, t test) for both PDX and patient tumor metastases datasets. For these three TFs, the associated patterns are represented across PDX and patient tumor metastasis compendium datasets: average differential expression of the TF-bound genes (red-blue heat maps), differential expression of the TF genes (yellow-blue heat maps), and differential levels of the TF gene siRNA signature (purple-cyan heat maps). A negative siRNA signature association indicates that knocking down the TF gene would result in a global pattern opposite TF gene overexpression.