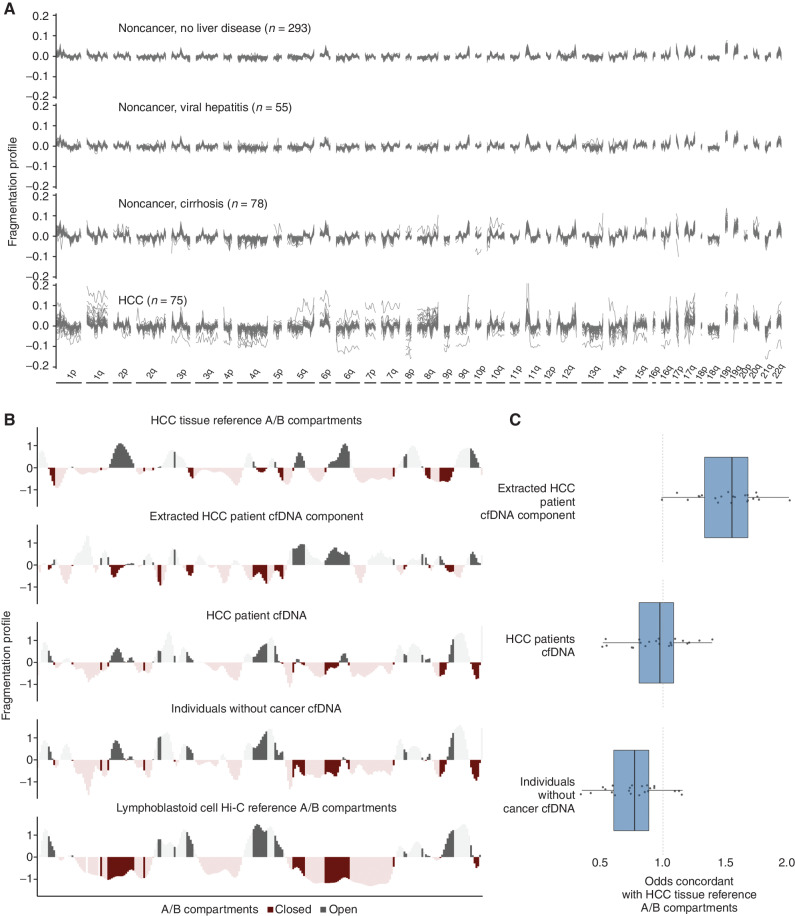
Figure 1.
Genome-wide fragmentation profiles reflect underlying chromatin structure. A, Fragmentation profiles of 501 individuals in 473 nonoverlapping 5-Mb genomic regions. Fragmentation profiles for individuals with cancer show marked heterogeneity as compared with noncancer individuals with and without liver disease. B, Comparison of plasma fragmentation features to reference A/B compartments. Track 1 shows A/B compartments extracted from liver cancer tissue (28). Track 2 shows a median liver cancer component extracted from the HCC plasma samples of 10 liver patients with high tumor fraction by ichorCNA (56). Track 3 shows the median fragmentation profile in the plasma for these 10 HCC samples, and track 4 shows the median profile for 10 healthy plasma samples. Track 5 shows A/B compartments for lymphoblast cells (28). These five tracks show chromosome 22 as an example, with darker shading indicating informative regions of the genome where the two reference tracks differ in domain (open/closed) or magnitude. C, Among these informative bins, for each chromosome, the log odds of the plasma component matching the HCC reference track in domain. Log odds greater than 1 indicate more similarity to the HCC reference track, whereas log odds less than 1 indicate more similarity to the lymphoblast reference track. The extracted HCC component has the greatest similarity to the HCC reference track, and the noncancer plasma has the greatest similarity to the lymphoblast reference track; the HCC plasma track is intermediate to the two.