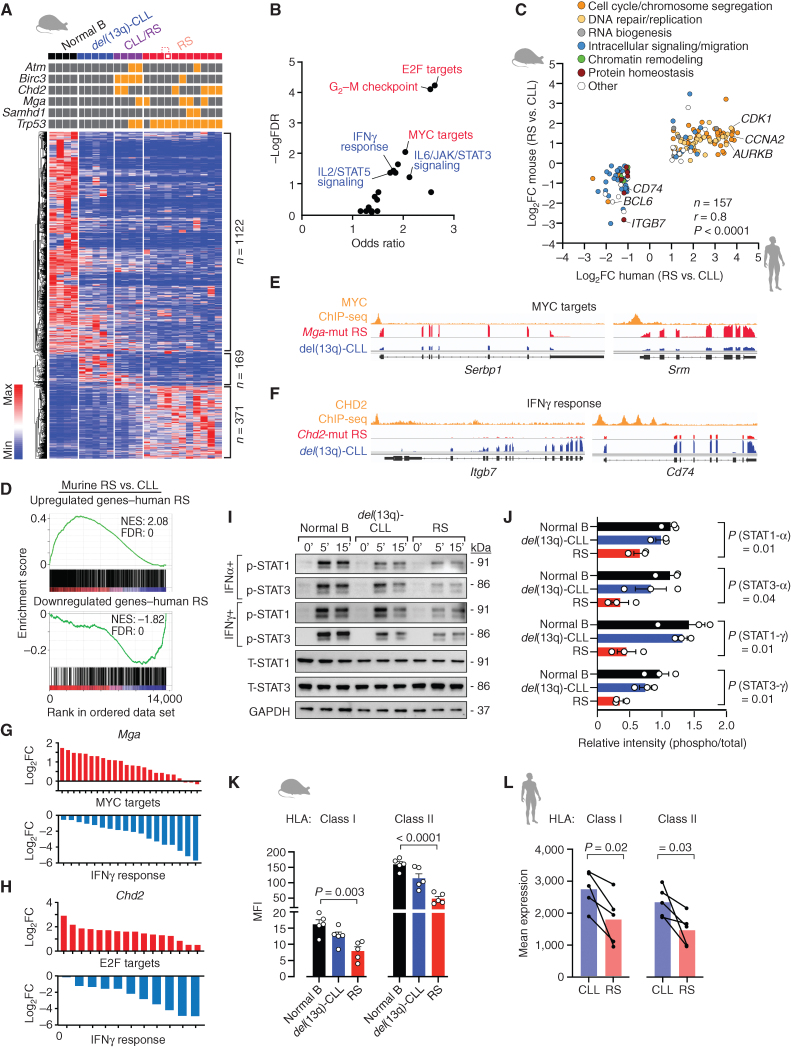
Figure 4.
Murine transcriptomes show strong similarity with human disease. A, Heat map of differentially expressed genes among the four different groups of mice (4 normal B, 5 del(13q)-CLL, 4 CLL/RS, and 11 RS). ANOVA FDR < 0.1 was used as a cutoff. B, Signaling pathways enriched for the differentially expressed genes in A. C, Concordantly modulated genes in human and murine RS and respective functional categories. P, Pearson correlation. D, Gene set enrichment plots showing the correlation of human RS versus CLL gene sets (upregulated, top; downregulated, bottom plot) in murine RS versus CLL data. NES, normalized enrichment score; FDR, false discovery rate. E, Representative IGV tracks of MYC ChIP-seq and RNA-seq data of two MYCMGA top targets in Mga-mutant RS and del(13q)-CLL. F, Representative IGV tracks of CHD2 ChIP-seq and RNA-seq data of two CHD2 top targets involved in the interferon γ pathway in Chd2-mutant RS and del(13q)-CLL. G, Log2FC [RS vs. del(13q)-CLL] of MGA targets included in MYC targets and IFNγ response categories. The top 500 MGA cistromeGO targets were used in the analysis. H, Log2FC [RS vs. del(13q)-CLL] of CHD2 targets included in E2F targets and IFNγ response categories. The top 500 CHD2 CistromeGO targets were used in the analysis. I, Immunoblot analysis of phospho-STAT1 and STAT3 (and respective totals), after 5- and 15-minute stimulation with soluble IFNα and IFNγ. One representative normal B-cell, del(13q)-CLL and RS sample is shown. GAPDH was probed, as control. J, Relative abundance of P-STAT1 and P-STAT3 levels across the three replicates of normal B cells, CLL and RS samples. Mean with SEM is displayed. P, ANOVA. K, Bar plot showing HLA class I and II MFI ratios (compared with isotype controls), as assessed by flow cytometry on normal B, del(13q)-CLL and RS cases (n = 5/group). Mean with SEM is displayed. P, ANOVA. L, Mean expression levels (in transcripts per million) of class I (HLA-A, -B, -C) and class II family (HLA-DR, -DQ) on RNA-seq data from 5 human CLL and matched RS cases. P, paired t test.