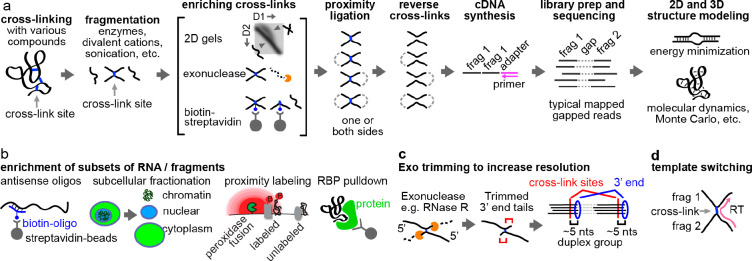
Figure 7.
General workflow of cross-link-ligation methods. (a) Basic pipeline. RNAs are cross-linked, fragmented, and enriched for the cross-linked fragments using various methods such as 2D gels, exonucleases, or streptavidin-beads for biotinylated cross-linkers. After proximity ligation and reversal of cross-links, adapters are ligated to the fragments for cDNA synthesis, which is followed by cDNA amplification and high throughput sequencing. The gapped reads provide spatial constraints for 2D and 3D structure modeling. (b) Specific subsets of RNAs can be enriched for structure analysis after cross-linking and before fragmentation, using various approaches, e.g., antisense oligos, subcellular fractionation, proximity labeling (biotinylation by the APEX system) and antibody pull down of specific RNA binding proteins. (c) Exonuclease trimming can be used to improve resolution. Exonucleases are blocked by monoadducts or cross-links, leaving a stub of fixed length, and the cross-link sites can be deduced by counting backward from the 3′ ends. (d) Template switching is an alternative of proximity ligation to capture the two fragments in a single read, based on the ability of the reverse transcriptase (RT) to switch templates as it encounters roadblocks.