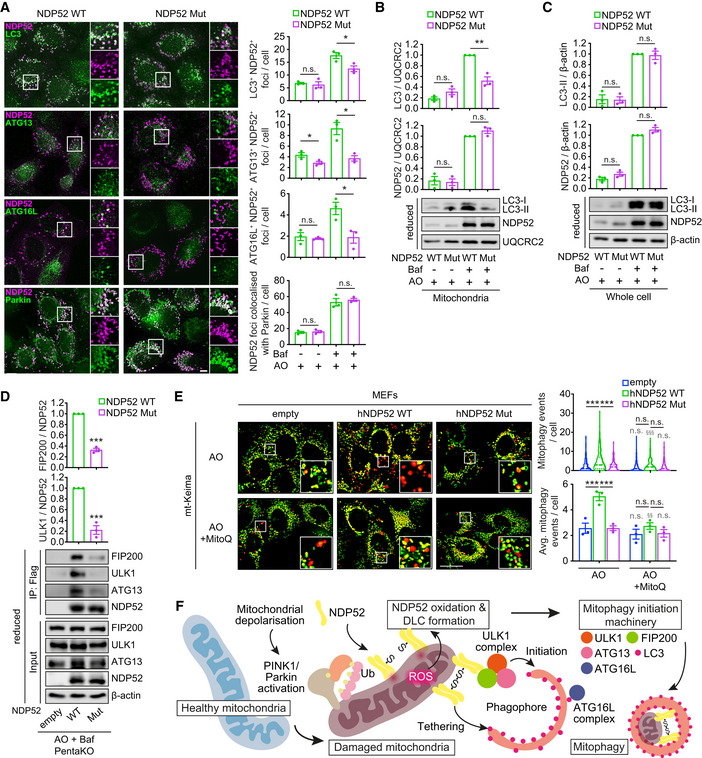
Figure 6. Oxidation of NDP52 facilitates the recruitment of autophagy proteins to damaged mitochondria.
-
A–DHeLa PentaKO + NDP52 WT or NDP52 Mut cells stably expressing YFP‐Parkin and mt‐mKeima were treated with 4 μM/10 μM AO for 2 h in the presence or absence of 400 nM Baf, followed by immunofluorescence analyses (A), immunoblotting for LC3 and NDP52 in mitochondrial fraction (B) or whole cell lysate (C) in reducing conditions, and co‐immunoprecipitation assay to analyse the interaction of NDP52 with FIP200, ULK1 and ATG13 (D). The number of foci of the indicated proteins colocalised with NDP52, or foci of NDP52 colocalised with Parkin, was quantified.
-
EMEFs stably expressing YFP‐Parkin, mt‐mKeima and empty, human NDP52 (hNDP52) WT or hNDP52 Mut were pre‐treated with or without 500 nM MitoQ for 21 h and treated with AO for 3 h followed by mitophagy measurement.
-
FSchematic representation of the mechanism of the damage‐induced mitophagy mediated by NDP52 DLC.
Data information: Data are mean ± s.e.m. (A–E) or displayed as cell popular violin plots (E). P values were calculated by one‐way ANOVA followed by Sidak test (for multiple groups) or unpaired two‐tailed Student's t‐test (for two groups) on three independent experiments (A–E). *, P < 0.05; **, P < 0.01; ***, P < 0.001; §§, P < 0.01; §§§, P < 0.001 (relative to MitoQ‐untreated condition); ns (non‐significant). Scale bars: 20 μm (A, E).
Source data are available online for this figure.
