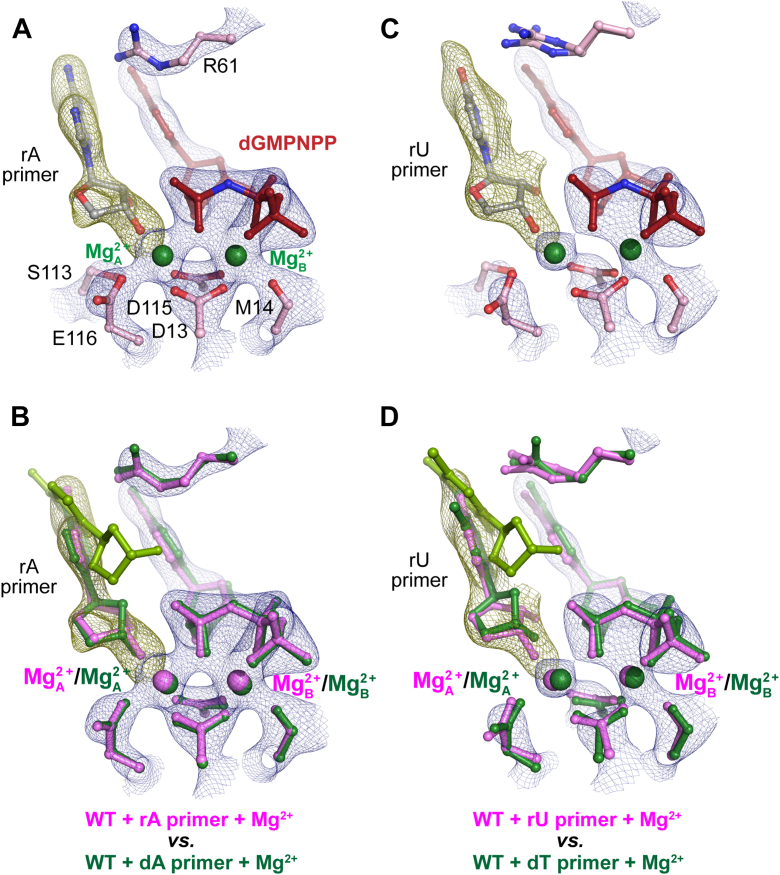
Figure 5.
Structural comparison between reactant states of incorrect nucleotide incorporation during dN or rN extension show increased inclination of primer terminus alignment during rA or rU extension misincorporation. A,C, structure of rA extension (A) and rU extension (C) WT Pol η mismatch reactant state complexed with Mg2+. B, D, structural overlay of dN (green) (PDB ID 4J9M and 4J9K) or rN (pink) extension structures with dGMPNPP:dT base-pair indicating differences in 3′-OH alignment. The misaligned primer terminus in the dN structure (green) is colored with yellow green. All electron density maps apply to the molecule colored in pink. The 2Fo-Fc map for everything including Me2+A and Me2+B, dGMPNPP, and catalytic residues and S113 (blue) was contoured at 2 σ. The Fo-Fc omit map for the primer terminus (green) was contoured at 3.5 σ in A, B, and 2.7 σ in C, D. rU, ribose uridine; dN, deoxynucleotide; Pol η, DNA polymerase η; rN, ribonucleotide; dT, deoxyribose thymine.