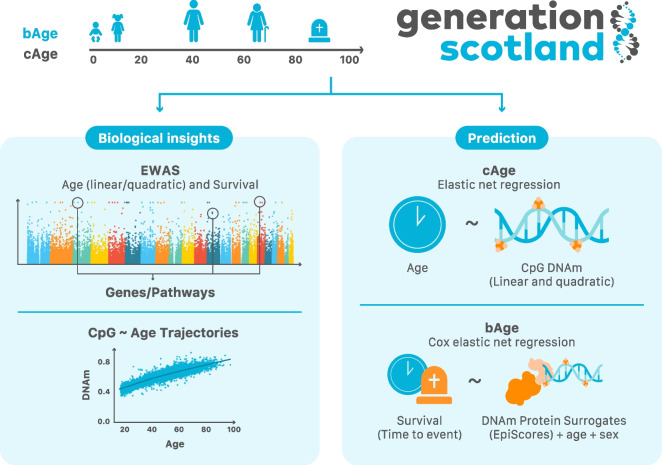
Fig. 1.
Study overview. Using the Generation Scotland cohort as our main data source, we explored the relationship between the epigenome and age/survival via EWAS, which also informed on genes of interest and potentially enriched pathways. We further characterised epigenome-wide CpG ~ age trajectories, which can be visualised in a new Shiny app, MethylBrowsR (https://shiny.igmm.ed.ac.uk/MethylBrowsR/). Finally, we refined epigenetic prediction of both cAge and bAge. Calculation of cAge can be performed either using a standalone script (https://github.com/elenabernabeu/cage_bage/tree/main/cage_predictor) or by uploading DNAm data to our MethylDetectR shiny app (https://shiny.igmm.ed.ac.uk/MethylDetectR/). As the weights for GrimAge and its component parts are not publicly available, bAge can only be calculated by using our standalone script (https://github.com/elenabernabeu/cage_bage/tree/main/bage_predictor), after obtaining GrimAge estimates from an external online calculator (http://dnamage.genetics.ucla.edu/new)