Fig. 1. Structures of Rutaceae and Meliaceae limonoids and proposed biosynthetic pathway.
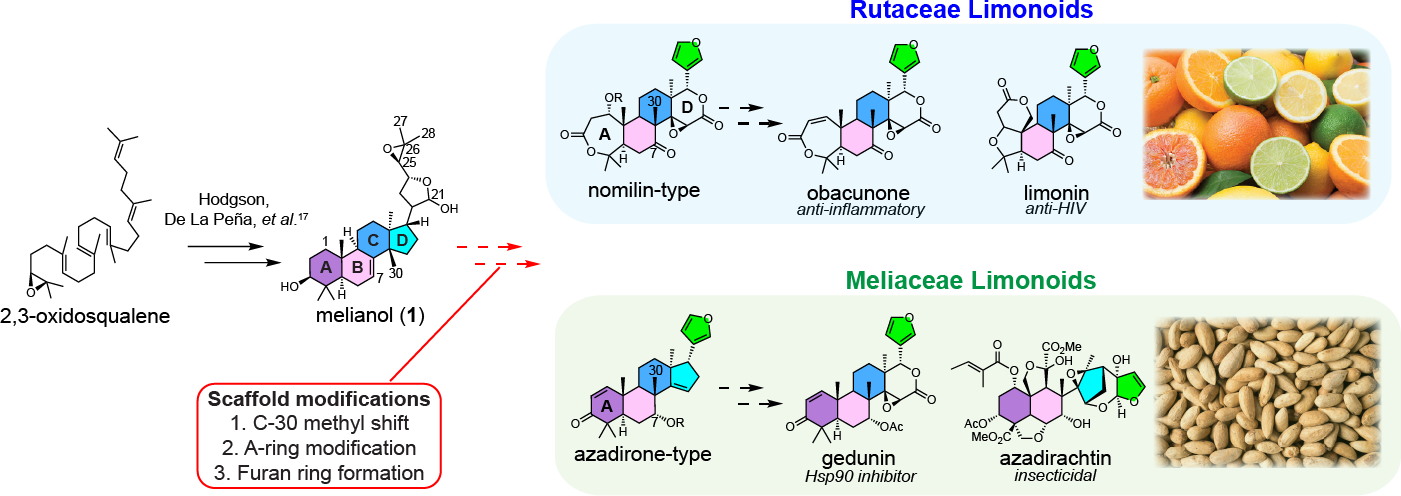
We previously characterized three conserved enzymes from both Citrus and Melia species that catalyze the formation of the protolimonoid melianol (1) from 2,3-oxidosqualene (20). Additionally, conserved scaffold modifications like C-30 methyl shift, furan-ring formation, and A-ring modification are proposed to convert protolimonoids to true limonoids. Beyond this, Rutaceae limonoids differ from Meliaceae limonoids in two key structural features: seco-A,D ring and C-7 modification, which are proposed to be the result of Rutaceae and Meliaceae specific modifications. Exceptions to this rule could potentially arise from late-stage species-specific tailoring (fig. S43). Rutaceae limonoids are derived from nomilin-type intermediates while Meliaceae limonoids are proposed to originate from azadirone-type intermediates. While the exact point of pathway divergence is unknown, comparative analysis of the various protolimonoid structures suggested that C-1, C-7, C-21 hydroxylation and/or acetoxylation are part of the conserved tailoring process. Obacunone and limonin are commonly found in various Citrus species (adapted photo by IgorDutina on iStock with standard license) and are responsible for the bitterness of their seeds. Azadirachtin (the most renowned Meliaceae limonoid) accumulates at high levels in the seeds of neem tree (photo by JIC photography), which are the source of commercial neem biopesticides.
