Fig. 2. Genomic and transcriptomic analysis of Citrus and Melia resources.
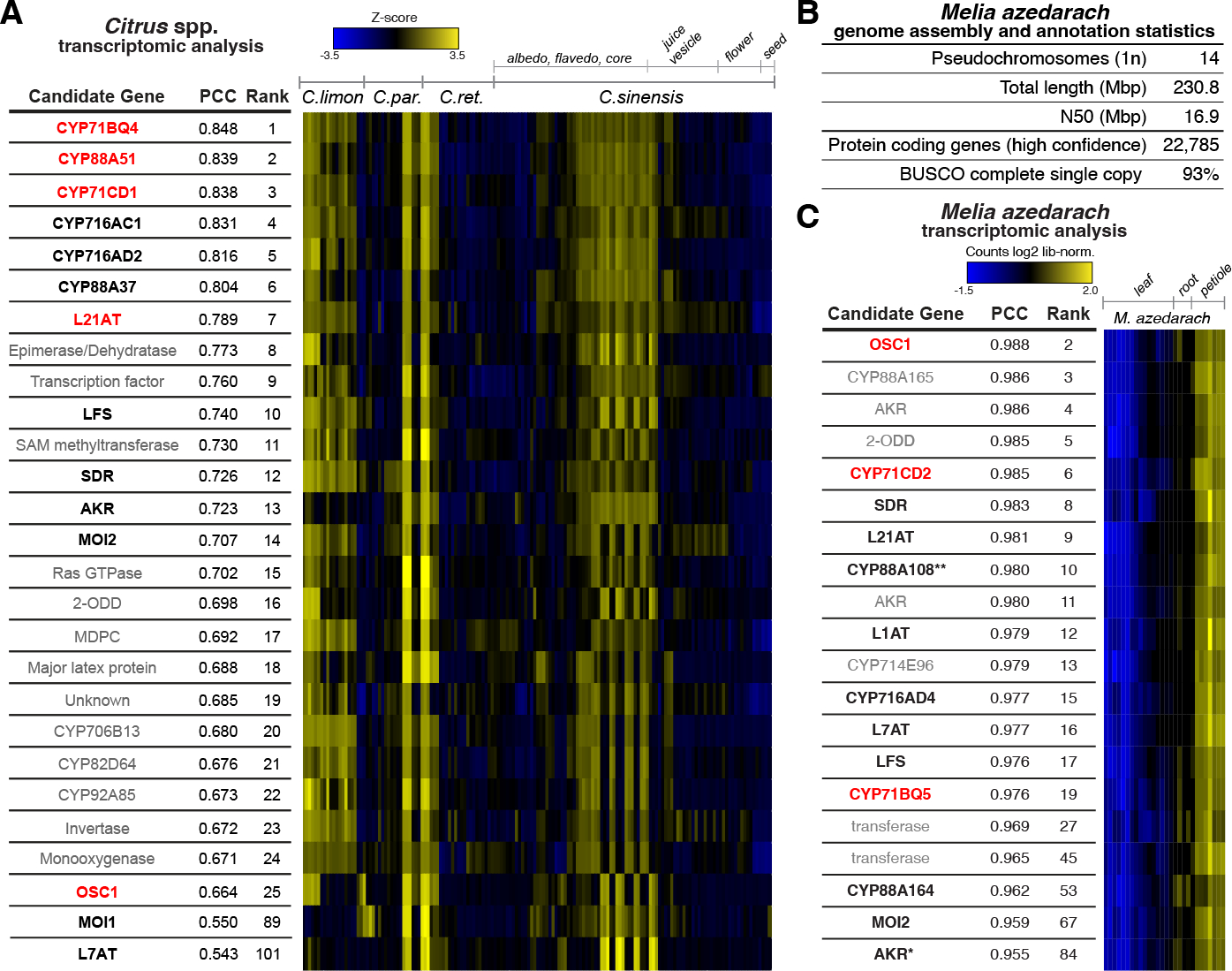
(A) Co-expression analysis of C. sinensis publicly available microarray expression data from NICCE (22) using CsOSC1, CsCYP71CD1, CsCYP71BQ4, CsCYP88A51 and CsL21AT as bait genes. Linear regression analysis was used to rank the top 25 genes based on Pearson’s correlation coefficient (PCC) to the bait genes of interest. Heat map displays Z-score calculated from log2 normalized expression across the fruit dataset. The reported PCC value corresponds to the average value calculated using each bait gene. Genes in red indicate bait genes used in analysis and genes in black are functional limonoid biosynthetic genes (table S18). Functional candidates outside of the top 25 genes are also included. For identification of individual bait genes used in this analysis see fig. S2. Enzymes have been abbreviated as follows: MOI = melianol oxide isomerase; CYP = cytochrome P450; L21AT = limonoid C-21-O-acetyltransferase; SDR = short-chain dehydrogenase; L1AT = limonoid C-1-O-acetyltransferase; L7AT = limonoid C-7-O-acetyltransferase; AKR = aldo-keto reductase; LFS = limonoid furan synthase; OSC = oxidosqualene cyclase.
(B) Summary of Melia azedarach pseudo-chromosome genome assembly and annotation statistics (fig. S3 to S4, table S1 to S2).
(C) Expression pattern of M. azedarach limonoid candidate genes selected based on PCC to melianol biosynthetic genes (MaOSC1, MaCYP71CD2 and MaCYP71BQ5 (20), shown in red) and biosynthetic annotation. Heatmap (constructed using Heatmap3 V1.1.1 (44), with scaling by row (gene)) includes genes that are ranked within the top 87 for co-expression and are annotated with one of six interpro domains of biosynthetic interest (IPR005123 (Oxoglutarate/iron-dependent dioxygenase), IPR020471 (Aldo/keto reductase), IPR002347 (Short-chain dehydrogenase/reductase SDR), IPR001128 (Cytochrome P450), IPR003480 (Transferase) and IPR007905 (Emopamil-binding protein)). Asterisks indicate the following: (*) full-length gene identified in transcriptomic rather than genomic data via sequence similarity to CsAKR ((table S10, table S19), (**) gene previously identified as homolog of limonoid co-expressed gene from A. indica (20)). Genes shown in black are newly identified functional limonoid biosynthetic genes (this study) (table S10).
