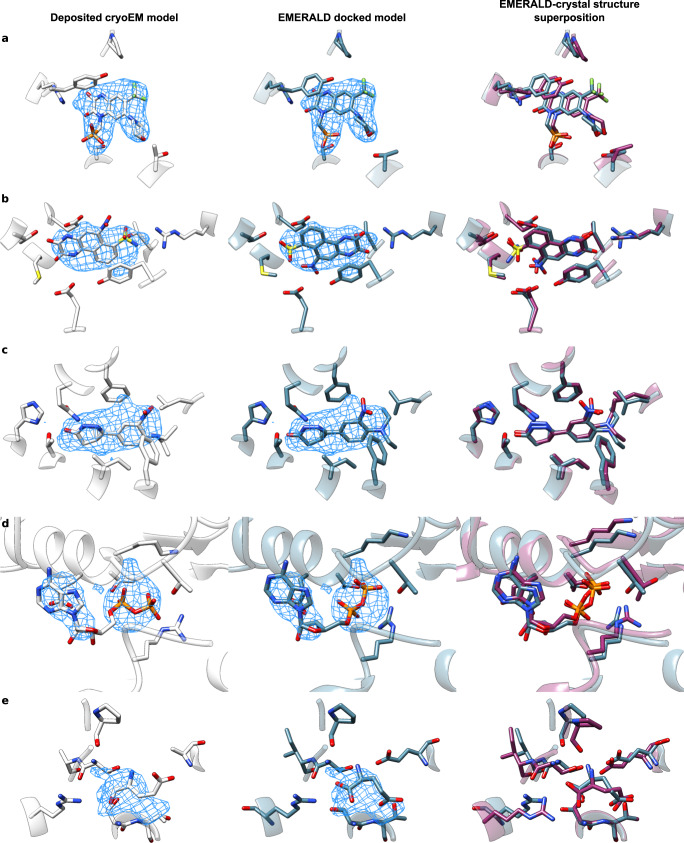
Fig. 3. Alternate conformations predicted with EMERALD match high-resolution crystal structures.
a–e Comparison of the deposited model (left, white), EMERALD model (center and right, blue), and higher resolution crystal model (right, purple). a Antagonist ZK200775 in AMPA receptor (EMDB: 23292, PDB: 7LEP, local resolution: 3.45 Å) and its associated crystal model (PDB: 5ZG2). b Molecule NBQX bound to the AMPA receptor (EMDB: 12805, PDB: 7OCE, local resolution: 2.75 Å) and its associated crystal model (PDB: 6FQH). c DNMDP bound to the SLFN12-PDE3A complex (EMDB: 23495, PDB: 7LRD, local resolution: 2.95 Å) and its associated crystal model (PDB: 7KWE). d ADP bound to ClpB (EMDB: 21553, PDB: 6W6E, local resolution: 4.42 Å) and its associated crystal model (PDB: 5LJ8). e Glutamate ligand in an AMPA receptor (EMDB: 12806, PDB: 7OCF, local resolution: 4.26 Å) and its associated crystal model (PDB: 3TKD).