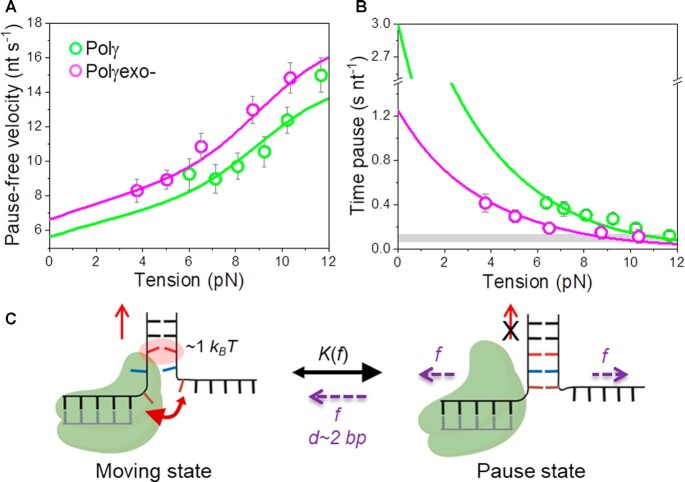
Figure 3.
Effect of tension on Polγ and Polγexo- strand displacement kinetics. For all plots: Polγ green symbols (N = 80), Polγexo- magenta symbols (N = 71). Error bars show standard errors. (A) For both holoenzymes pause-free velocities (nt s−1) increased with tension continuously towards values measured during primer extension (Figure S1C). Green and magenta lines are the fits of the strand displacement model (SI) to Polγ and Polγexo- data, respectively. (B) Tension dependencies of the average residence times at the pause state per nucleotide (Tp(f), s nt−1). Green and magenta lines are the fits to Polγ and Polγexo- data, respectively, with a two state model (Eq. 1). Grey box show average Tp(f) values measured under primer extension conditions in the absence of mtSSB, Figure S1D and (16). (C) Diagram illustrating the two-state model in which the holoenzyme alternates between moving and pause or non-productive states during the strand displacement reaction. In the moving state, two template nucleotides (brown and blue) are bound to the pol site and the holoenzyme advances through the dsDNA destabilizing partially the first base pair of the junction (in red) with interaction energy of ΔGint ∼1 kBT per dNTP incorporation step. In the absence of tension, the regression pressure of the dsDNA fork outcompetes the holoenzyme for the template (two headed arrow), which shifts the equilibrium towards the pause or non-productive state strongly (K(0) >1, Table 1) and restricts the probability of finding Polγ and Polγexo- in the moving state to ∼4 and 12%, respectively (SI). Application of tension (f) to the hairpin decreases the rewinding kinetics and/or favors the unwinding of first ∼2 bp of the fork (d), which shifts the equilibrium towards the moving state.