Figure 4.
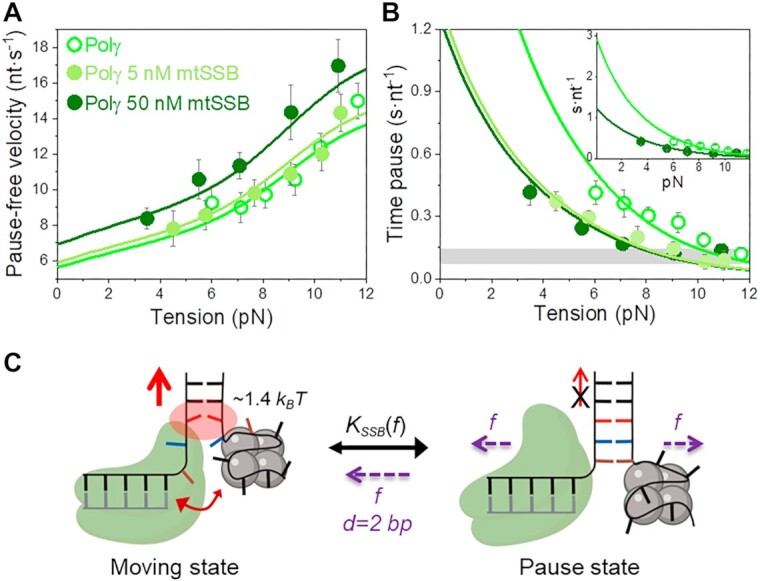
Effect of mtSSB on the tension dependent strand displacement kinetics of Polγ. (A) 50 nM (N = 40) but not 5 nM (N = 20) mtSSB stimulated the pause-free velocity of Polγ at all tensions. Green lines correspond to the fits of the strand displacement model to data in the absence and presence of mtSSB. (B) 5 and 50 nM concentrations of mtSSB decreased average residence time at pause state per nucleotide (Tp(f), s nt−1) of Polγ at all tensions. Grey box shows average Tp(f) values obtained under primer extension conditions in the absence of mtSSB (16). Green lines are the fits of two-state model (Eq. 1) to data in the absence and presence of mtSSB. mtSSB binding to the displaced strand decreases ∼2–3 times the average residence time of Polγ at a pause or non-productive state. For both figures error bars show standard errors. Inset shows the intersection of the fits with the Y-axis. (C) Diagram illustrating the two-state model in the presence of mtSSB. Polγ alternates between moving and pause or non-productive states. In the moving state, two template nucleotides (brown and blue) are bound at the pol active site and the holoenzyme-mtSSB complex destabilizes partially the first base pair of the DNA hairpin with interaction energy a ∼40% higher than in the absence of mtSSB (ΔGint ∼1.4 kBT per dNTP incorporated). In addition, mtSSB decreases the fork regression kinetics (represented by a two-headed arrow), which in turn, increases the probability of finding the holoenzyme at the moving state from ∼4 to ∼12% (SI). Even in the presence of mtSSB, the equilibrium is shifted towards the pause or inactive-state (KSSB(0) > 1, Table 1). Destabilization of ∼2 base pairs (d) of the DNA junction by application of mechanical tension (f) is required to shift the equilibrium towards the moving state.
