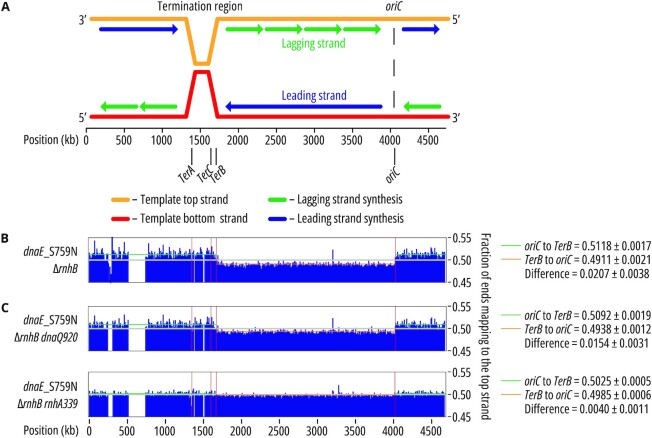
Figure 4.
Genome-wide mapping of pol III strand-specific ribonucleotide incorporation. (A) A schematic representation of E. coli chromosome replication. oriC, the replication origin, as well as the termination region containing TerA, TerB and TerC are marked. (B, C) Results for pol IIIα_S759N active site mutant strains are shown with the fraction of end-mapped reads in bins of 10 000 bp after background normalization using the wild-type strain. The numbers depicted on the right side of the graphs show vertical viewing range. Average fraction of ends mapping to the top strand ± SD between oriC and TerB (green), or TerB and oriC (orange), is presented next to the figure. Results for two other dnaE mutants (dnaE_S759C and dnaE_S759T) are shown in Supplementary Figure S4A. (B) The dnaE_S759N mutant shows a strong strand switch that correlates to the positions of oriC and TerB. The average difference between the leading and lagging strands is 0.0207. Three biological replicates for each dnaE allele are presented in Supplementary Figure S3A. (C) The strand bias in the strain expressing dnaE_S759N mutant slightly decreases when proofreading is impaired (dnaQ920) to 0.0154 and is essentially abolished (0.0040) when in combination with an inactive RNase HI (rnhA339::cat). Three biological replicates of each strain are presented in Supplementary Figure S3B, C.