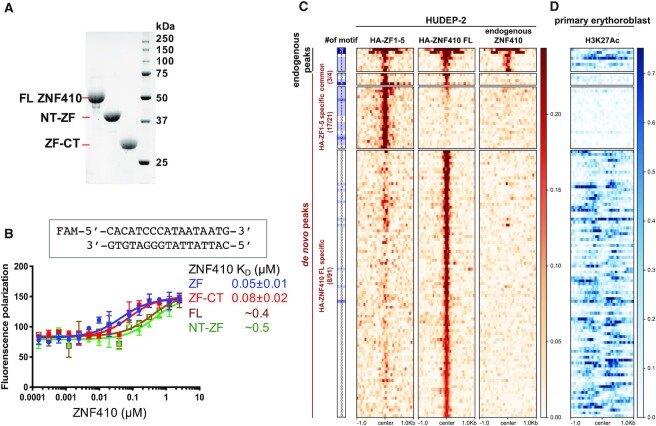
Figure 2.
ZF1-5 has stronger DNA binding in the absence of the N-terminal domain. (A) An 18% SDS-PAGE gel of Bio-RAD stain-free imaging showing the recombinant protein samples used in this study. (B) Fluorescence polarization assay of recombinant ZNF410 protein segments with and without N- or C-terminal additions. Data represent the mean ± SD of three independent determinations (N = 3). (C) Unique binding sites of HA-ZF1-5 and HA-ZNF410 FL in HUDEP-2 cells. Heatmaps of the signal intensities of the HA-ZF1-5 (left column), HA-ZNF410 FL (middle) and endogenous ZNF410 (right) ChIP-seq peaks in HUDEP-2 cells (GSE154960). At the far left is an indication of binding motif occurrence. (D) The corresponding H3K27ac mark in primary human erythroblasts. Note that HUDEP-2 cells exhibit similar gene expression patterns to human HSPC-derived erythroid cells (primary erythroblasts) (59).