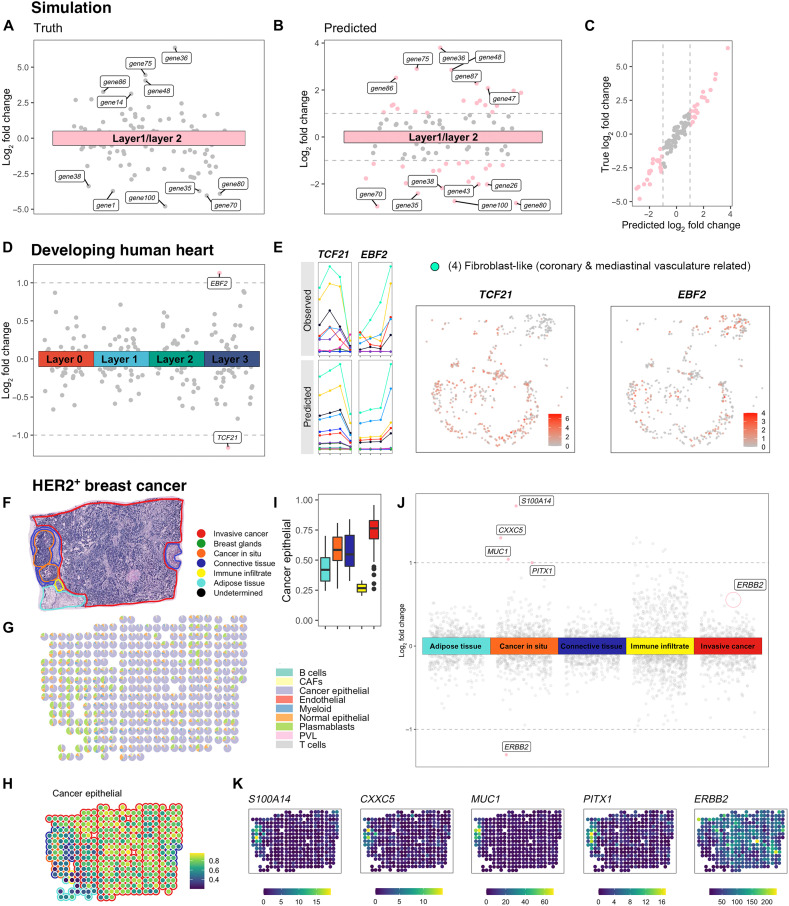
Fig. 3. LDE genes identified by POLARIS on three datasets.
On simulated data: (A) True log2 fold change of mean gene expression, layer 1 over layer 2. (B) POLARIS-inferred log2 fold change of mean gene expression, layer 1 over layer 2. POLARIS-identified LDE genes are colored as pink, otherwise gray. (C) POLARIS-inferred log2 fold change of gene expression across layers achieves high correlation (Pearson’s correlation = 0.974) with the true log2 fold change. On developing human heart data: (D) POLARIS-inferred log2 fold change of gene expression across layers. POLARIS-identified LDE genes are colored as pink, otherwise gray. (E) Left: Observed mean gene expression in scRNA-seq data (top) and POLARIS-inferred gene expression location parameter (bottom) of each cell type across layers. Lines are colored by cell types. X axis indicates layer status: From left to right is layer 0, 1, 2, and 3. Right: Gene expression of TCF21 and EBF2 in the fibroblast-like (coronary and mediastinal vasculature related) cells. On HER2+ breast cancer data: (F) Pathologist annotation on slide A1. (G) POLARIS-inferred cell composition. (H) POLARIS-inferred cancer epithelial cell proportion. (I) Distribution of POLARIS-inferred cancer epithelial cell proportions in each layer [color scheme is the same as in (E)]. (J) POLARIS-inferred log2 fold change of gene expression across layers. Points with absolute value greater than 1 are colored as pink, otherwise gray. (K) Gene expression profiles of POLARIS-identified LDE genes in the cancer in situ layer.