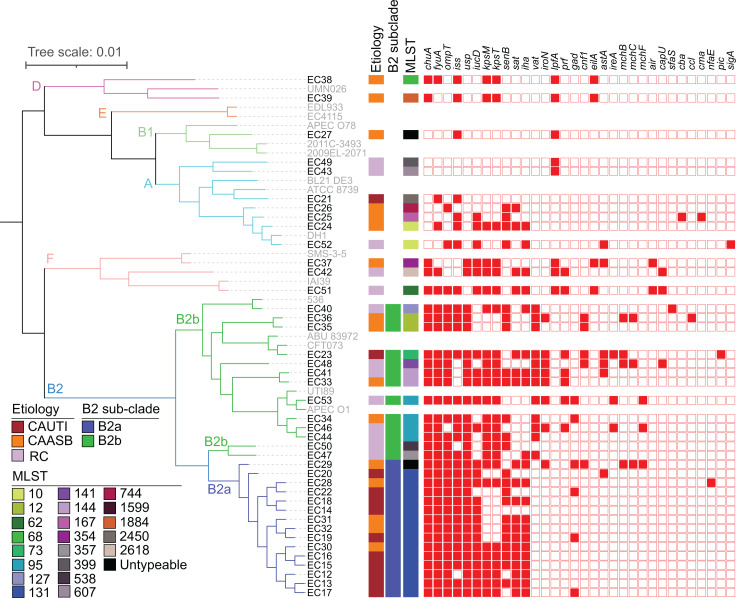
Figure 1. Phylogenetic distribution of 41 clinical E. coli isolates.
Core-genome alignment was constructed into a maximum likelihood tree with raxML and viewed in iTOL, with 6 phylotypes identified. In silico multilocus sequence types (MLST) were identified using BLASTN to the E. coli MLST database, with 21 STs identified. Virulence factors (VFs) were annotated in the E. coli draft genomes using VirulenceFinder v1.5 and blastp to previously described genes, with 32 VFs identified. The strain names of E. coli sequenced in this study were in black and reference E. coli strains were in gray.