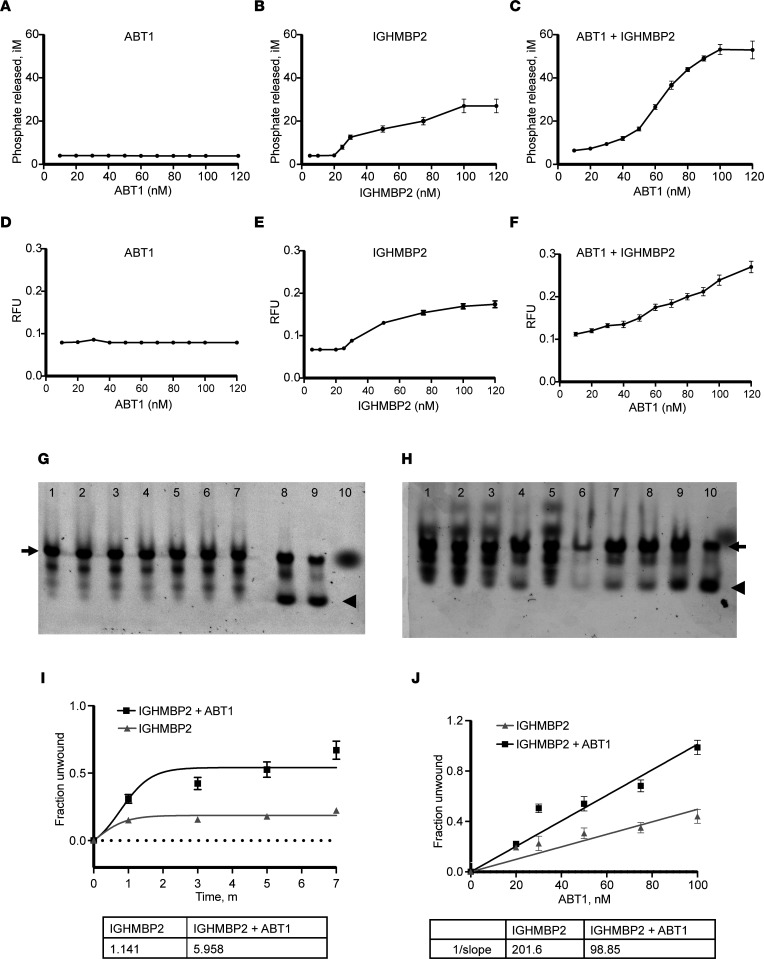
Figure 4. The association of ABT1 with IGHMBP2 increases the ATPase and helicase activity as well as the processivity of IGHMBP2.
(A) ATPase activity measured with increasing concentrations of ABT1. (B) Increasing concentrations of IGHMBP2. (C) IGHMBP2 (100 nM) incubated with increasing concentrations of ABT1. The data represent mean values from 3 independent experiments and 9 readings. (D) Helicase activity measured with increasing concentrations of ABT1. (E) Increasing concentrations of IGHMBP2. (F) IGHMBP2 (100 nM) incubated with increasing concentrations of ABT1. The data represent mean values from 3 independent experiments and 9 readings. (G) Lanes 1–4 show unwinding analyses of TP31-18mer incubated with 100 nM IGHMBP2 after 1, 3, 5 and 7 minutes, respectively. Lanes 5–8 show unwinding analysis of TP31-18mer incubated with 100 nM IGHMBP2 + 100 nM ABT1 after 1, 3, 5, and 7 minutes, respectively. Lane 9 shows heat-induced TP31-18mer DNA separation (95°C for 5 seconds) used as a positive control. Lane 10 shows Cy3 labeled DNA TP31-18mer alone (negative control). (H) Lanes 1–4 show unwinding analysis of TP31-18mer incubated with 100 nM IGHMBP2 after 1, 3, 5, and 7 minutes, respectively. Lane 5 shows unwinding analysis of TP31-18mer incubated with 100 nM IGHMBP2 + 20 nM ABT1 with no incubation. Lanes 6–10 show unwinding analysis of TP31-18mer with 100 nM IGHMBP2 + increasing concentrations of ABT1 (20, 40, 50, 75, 100 nM) after 7 minutes of incubation. (I) Quantification of 3 independent experiments of unwinding analyses as shown in G. (J) Quantification of 3 independent experiments of unwinding analyses as shown in H. Arrow indicates the double-stranded duplex, and the arrowhead indicates the resolved duplex. DNA substrate was used at a final concentration of 10 nM. iM, free phosphate produced in μM; RFU, relative fluorescence units. Data are represented as mean ± SD; 1-tailed paired t test was used to calculate statistical significance.