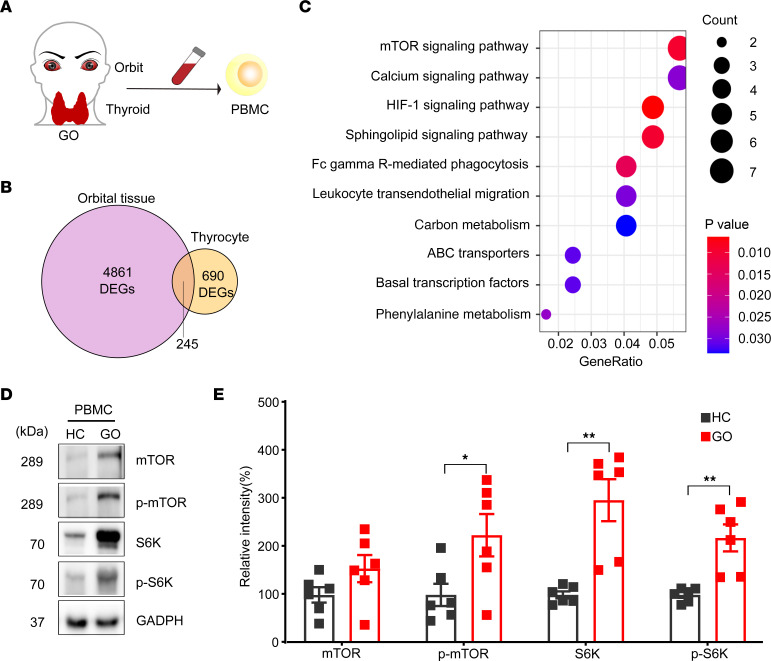
Figure 4. The mTORC1 signaling pathway is upregulated in the orbital tissue, thyrocytes, and PBMCs of patients with GO.
(A) Orbits, thyroids, and PBMCs from patients with GO were all investigated to determine the key pathway involved in GO pathogenesis. (B) A Venn diagram of differentially expressed genes (DEGs) in the GO gene set identified in the GSE58331 data set (orbital tissues; GO vs. HC) and in the GSE9340 data set (thyrocytes; GO vs. GD). A total of 245 GO-specific genes were identified. (C) Top 10 enrichment signatures revealed by KEGG analysis of the 245 GO-specific genes. The mTOR signaling pathway was the top enriched pathway in GO. P values are indicated by color. The number of DEGs in enrichment signatures were indicated by circle size. (D) Representative images of Western blot, which analyzed the key signaling molecules involved in mTORC1 signaling pathways from PBMCs of HCs and patients with GO. (E) Bar plots for the relative intensities in the Western blot of mTOR, p-mTOR, S6K, and p-S6K in the PBMCs of healthy controls and patients with GO (n = 6 for each group). Values represent the mean ± SEM. *P < 0.05 and **P < 0.01, by 2-tailed, unpaired Mann-Whitney-Wilcoxon rank test and independent-sample 2-tailed t tests for E.