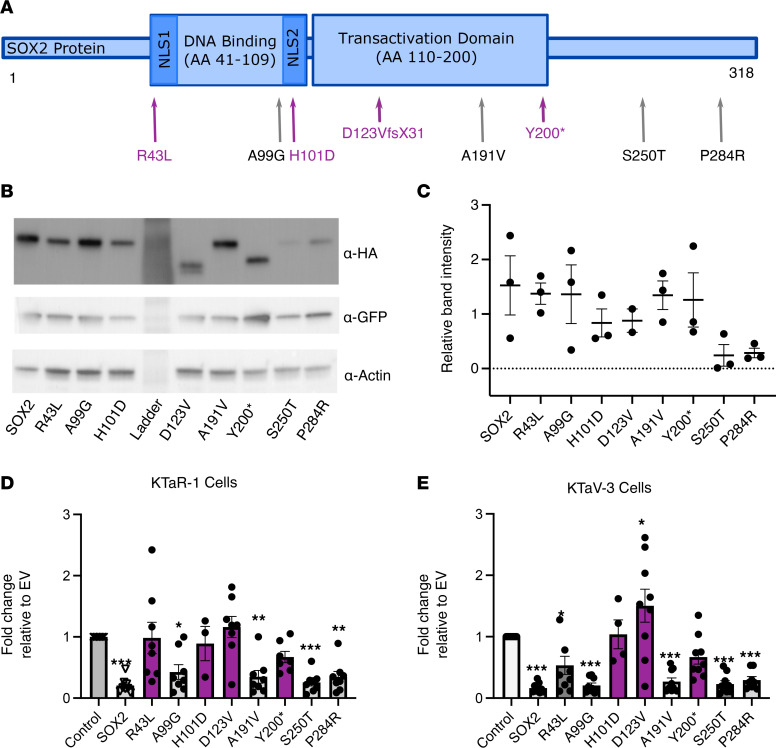
Figure 4. Mutations in SOX2 interfere with SOX2-mediated repression of KISS1 transcription.
(A) Schematic of the SOX2 protein with patient mutation locations indicated. Protein mutation nomenclature follows recommendations by the Human Genome Variation Society. NLS, nuclear localization signal. (B) Western blot of KTaR-1 cells transfected with SOX2-HA WT or a mutation-harboring hSOX2 plasmid. (C) Quantification of Western blot in B. Total volume for each band normalized to remove background and displayed relative the transfection control, GFP. Actin used as a loading control. N = 3. (D and E) hKiss-luc was cotransfected with SOX2, a mutation-harboring variant of SOX2, or an empty vector (EV) into KTaR-1 or KTaV-3 cells, respectively. SOX2 protein represses expression from hKiss-luc by 79% in KTaR-1 and 84% in KTaV-3 cells. Mutant SOX2 proteins that did not repress the expression from hKiss-luc are indicated in purple in A, D, and E. Values were normalized relative to EV. N = 3–9. Data represent mean ± SEM. Data were analyzed using 1-way ANOVA with Dunnett’s multiple-comparison post hoc test if ANOVA was significant. Significance indicated by *P < 0.05, **P < 0.01, ***P < 0.001.