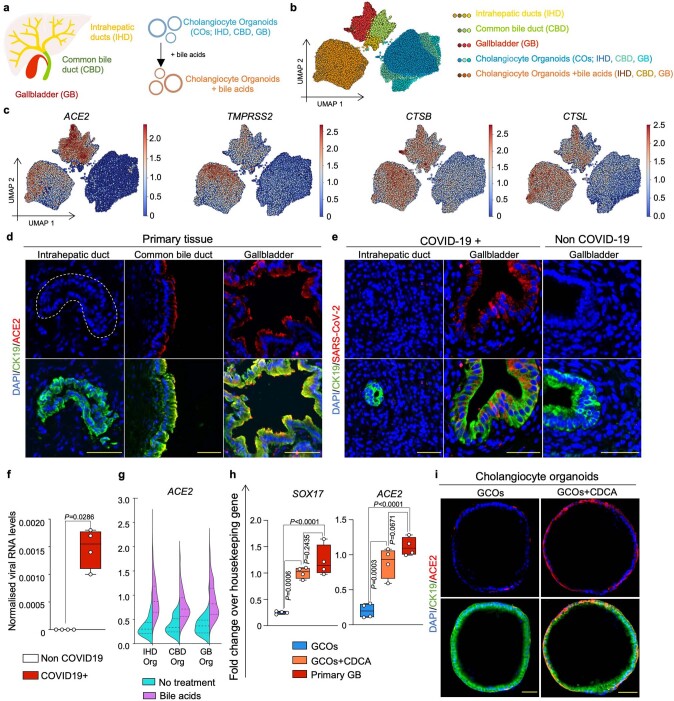
Extended Data Fig. 1. Expression of SARS-CoV-2 entry genes in cholangiocytes.
(a) Schematic illustration of different primary human cholangiocyte populations corresponding to different areas of the biliary tree and COs derived from different areas of the biliary tree grown in absence or presence of the bile acids. (b) UMAP plot illustrating different cholangiocyte populations from (a) analysed by scRNA-seq. (c) UMAP plots showing that viral entry related genes are predominantly expressed in extrahepatic cholangiocytes and COs treated with bile acids. (d–e) Immunofluorescence illustrating that ACE2 is expressed in extrahepatic cholangiocytes (d) and that SARS-CoV-2 infects gall bladder cholangiocytes of patients with COVID-19 but not intrahepatic cholangiocytes (e). N = 4 independent samples. Scale bars 50 μm. (f) QPCR confirming detection of SARS-CoV-2 RNA in bile of patients with COVID-19. Housekeeping gene, HMBS; n = 4; two-tailed Mann–Whitney test; centre line, median; box, interquartile range (IQR); whiskers, range; bars, standard deviation. (g) Violin plot of scRNA-seq data from (b) showing that COs upregulate ACE2 when treated with bile acids regardless of their region of origin. (h) QPCR validating that upon treatment with the bile acid CDCA COs assume a gall bladder identity expressing the gall bladder marker SOX17 and upregulating ACE2 at levels comparable to primary gall bladder. Housekeeping gene, HMBS; n = 4 independent experiments; one-way ANOVA adjusted for multiple comparisons; ns, non-significant; centre line, median; box, interquartile range (IQR); whiskers, range; bars, standard deviation. (i) Immunofluorescence showing that CDCA induces ACE2 expression in GCOs. N = 4 independent experiments. Scale bars 50 μm.