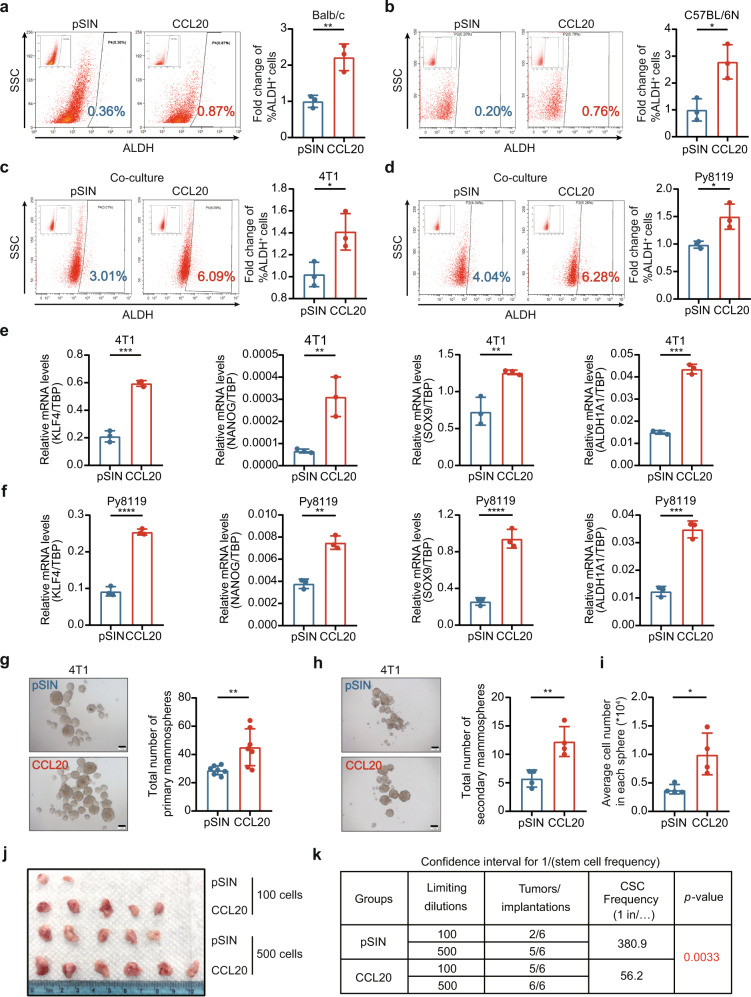
Fig. 3.
CCL20-modulated PMN-MDSCs enhanced the stemness of breast cancer cells. Balb/c mice or C57BL/6N mice were orthotopically transplanted with pSIN-/CCL20-overexpressing 4T1 or Py8119 cells at the fourth mammary fat pads, respectively. a, b The percentage of ALDH+ BCSCs was determined by ALDEFLUOR assay in tumor cells (CD45−CD140b−CD31−) from tumors of Balb/c mice (a) or C57BL/6N mice (b). Bar graph was presented as the mean of three biologically independent experiments (mean ± SEM). c, d PMN-MDSCs, sorted from pSIN-/CCL20-overexpressing 4T1 or Py8119 cell allograft tumors, were labeled as pSINPMN or CCL20PMN in Figures and as pSIN-PMN-MDSCs or CCL20-modulated PMN-MDSCs in main text. 4T1 or Py8119 cells were co-cultured with pSIN-/CCL20-modulated PMN-MDSCs for 3 days. The percentage of ALDH+ BCSCs was determined by ALDEFLUOR assay in 4T1 (c) or Py8119 cells (d). Bar graph was presented as the mean of three biologically independent experiments (mean ± SEM). e, f 4T1 or Py8119 cells were co-cultured with pSIN-/CCL20-modulated PMN-MDSCs for 3 days. The mRNA expression levels of several stemness-related genes were analyzed in 4T1 (e) or Py8119 cells (f) by qRT-PCR. Bar graph was presented as the mean of three biologically independent experiments (mean ± SEM). g–i Self-renewal capability was determined by forming both primary mammospheres (g, n = 7 for each group) and secondary mammospheres (h, n = 4 for each group) in 4T1 cells mix-cultured with pSIN-/CCL20-modulated PMN-MDSCs. The average cell number of each secondary mammosphere was shown (i), and bar graph was presented as mean ± SEM. Scale bar, 1 mm. j, k 4T1 cells were mix-cultured with pSIN-/CCL20-modulated PMN-MDSCs for 3 days, and then sorted by flow cytometry and engrafted to the fourth mammary fat pads of Balb/c mice at a limited dilution (n = 3 for each group, two injection sites each mouse, 100 or 500 cells/site). Tumor image was shown (j). The stem cell frequency in tumor tissue was calculated by the limited dilution assay (k). The stem cell frequency and p-value calculation were based on the positive tumor sites. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001