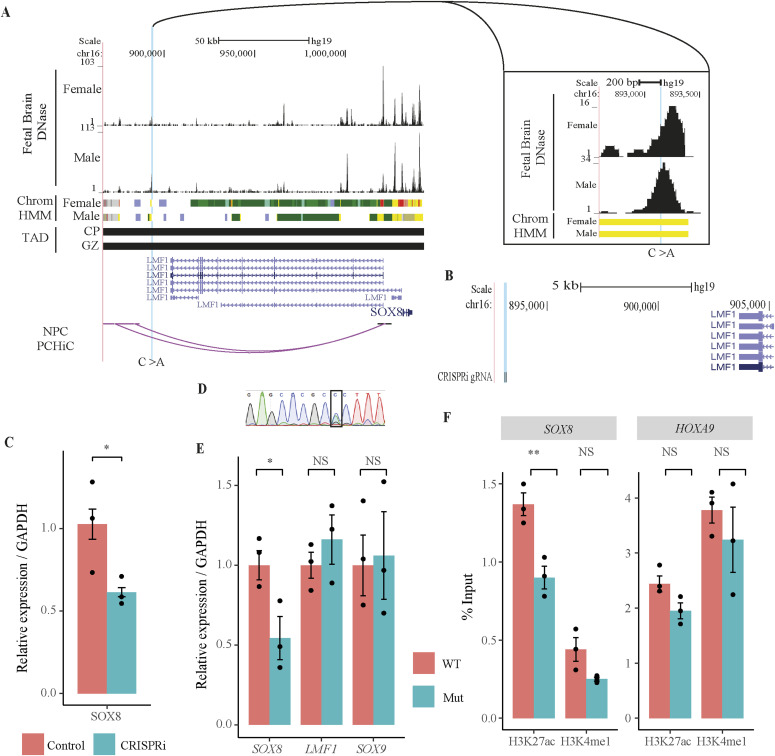
Figure 4. Characterisation of SOX8 enhancer DNM using CRISPR.
(A) USCS tracks depicting male and female fetal brain DNase hypersensitivity peaks, ChromHMM tracks, fetal brain topologically associated domains, and enhancer–promoter interactions. Yellow bars in chromHMM tracks indicate enhancers. (B) USCS tracks depicting the location of CRISPRi gRNA with respect to the 3′ end of the LMF1 gene. (C) Relative expression levels of gene SOX8 in neuronal progenitor cells with the CRISPRi of SOX8 enhancer and non-target guide RNA controls, normalised to GAPDH. (D) Sanger sequencing trace file shows a heterozygous knock-in mutation in HEK293T cells using CRISPR/Cas9-mediated homology-directed repair. The black box highlights the location of the DNM. (E) Relative expression levels of SOX8, LMF1, and SOX9 in WT and mutant cell line normalised to GAPDH. (F) Percentage of input (% input) of H3K27ac and H3K4me1 levels at SOX8 enhancer and HOXA9 promoter (control region) in WT and mutant cell lines. The significance level was calculated using a two-tailed t test. *** indicates P-value ≤ 0.001, ** indicate P-value between 0.01 and 0.001, whereas * indicates P-value between 0.01 to 0.05. WT, wild type; Mut, mutant.