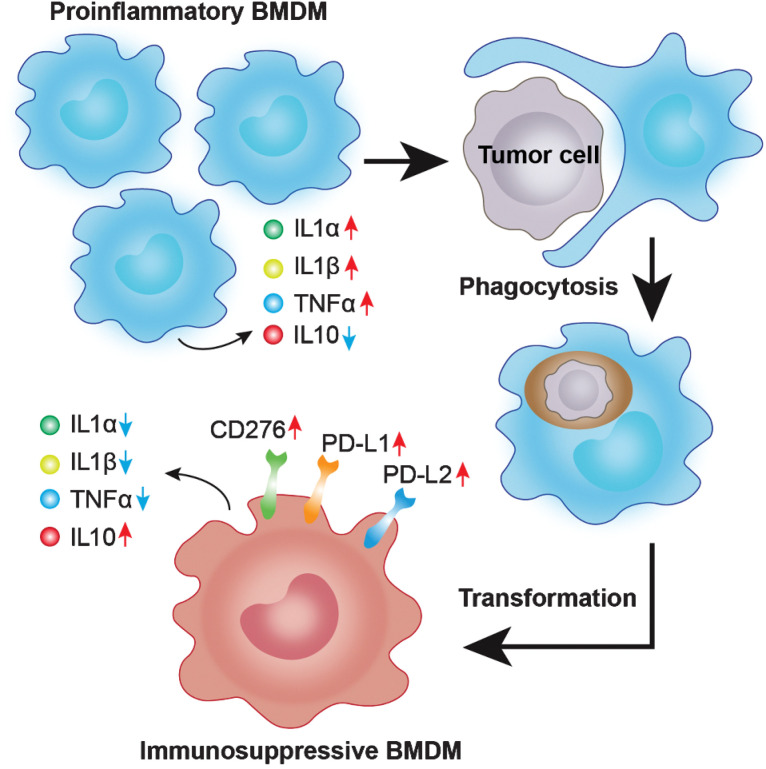
Bone marrow–derived macrophages phagocytose glioblastoma cells to form double-positive cells, dually expressing macrophage and tumor signatures that are transformed into M2-like macrophages and drive immunosuppression.
Abstract
Tumor-associated macrophages (TAM) play a crucial role in immunosuppression. However, how TAMs are transformed into immunosuppressive phenotypes and influence the tumor microenvironment (TME) is not fully understood. Here, we utilized single-cell RNA sequencing and whole-exome sequencing data of glioblastoma (GBM) tissues and identified a subset of TAMs dually expressing macrophage and tumor signatures, which were termed double-positive TAMs. Double-positive TAMs tended to be bone marrow–derived macrophages (BMDM) and were characterized by immunosuppressive phenotypes. Phagocytosis of glioma cells by BMDMs in vitro generated double-positive TAMs with similar immunosuppressive phenotypes to double-positive TAMs in the GBM TME of patients. The double-positive TAMs were transformed into M2-like macrophages and drove immunosuppression by expressing immune-checkpoint proteins CD276, PD-L1, and PD-L2 and suppressing the proliferation of activated T cells. Together, glioma cell phagocytosis by BMDMs in the TME leads to the formation of double-positive TAMs with enhanced immunosuppressive phenotypes, shedding light on the processes driving TAM-mediated immunosuppression in GBM.
Significance:
Bone marrow–derived macrophages phagocytose glioblastoma cells to form double-positive cells, dually expressing macrophage and tumor signatures that are transformed into M2-like macrophages and drive immunosuppression.
Graphical Abstract
Introduction
Glioblastoma (GBM), the most common and malignant primary brain tumor in adults, has a dismal prognosis, with a median survival of 15 months (1). Although immunotherapy has substantially improved the clinical outcomes of patients across multiple tumor types, such as advanced-stage melanoma (2) and non–small cell lung cancer (3), the highly immunosuppressive microenvironment in GBM patients hampers the effectiveness of immunotherapy (4, 5).
Accumulating evidence suggests that tumor-associated macrophages (TAM) mediate systemic immunosuppression in GBM (6). TAMs, the primary component of the GBM tumor microenvironment (TME), are ontogenetically composed of two populations: the brain-resident microglia (MG), generated from embryonic yolk sac precursors and maintained via prolonged longevity and self-renewal (7), and the bone marrow–derived macrophages (BMDM), derived from circulating monocytes and recruited to the TME in the context of brain malignancies (7). Emerging studies reveal the immunosuppressive roles of BMDMs in GBM. Müller and colleagues reported that BMDMs tend to express immunosuppressive cytokines compared with MG (8). Pinton and colleagues also demonstrated that the immunosuppressive microenvironment of gliomas depends on the accumulation of BMDMs (9). Nevertheless, how BMDMs are transformed into an immunosuppressive phenotype in the GBM TME remains unclear.
Several studies have described the existence of cells that coexpress TAM markers and tumor cell markers (10, 11). Double-positive cells are formed by the hybridization of TAMs and GBM cells (12). The hybrids exhibit unique transcriptome profiles via nuclear reprogramming and contribute to GBM invasion. Another report (11) stated that the double-positive cells are formed by the fusion of neoplastic cells and macrophages, and the fusion cells contribute to tumor heterogeneity and metastasis. However, few studies have investigated the immune regulatory functions of the double-positive cells in the GBM TME.
To address these problems, we utilized single-cell RNA sequencing (scRNA-seq) and whole-exome sequencing (WES) data of GBM tissues and identified a subset of TAMs that expressed macrophage and tumor signatures simultaneously, which were termed double-positive TAMs and tended to be BMDM-like. We found that the double-positive TAMs were formed through phagocytosis of glioma cells by BMDMs and exhibited enhanced immunosuppressive activities.
Materials and Methods
Human GBM specimens
Human GBM specimens were collected from patients who were pathologically diagnosed with GBM at the First Affiliated Hospital of Nanjing Medical University. All the patients involved provided written informed consent. All the sampling steps in this study were conducted in accordance with the recognized ethical guidelines of the Declaration of Helsinki and approved by the ethics committee of the First Affiliated Hospital of Nanjing Medical University. The clinical information of GBM patients was listed in Supplementary Table S1.
Cell culture
Glioma stem cells (GSC; WL1), normal neural stem cells (NSC; HNP1), and THP1 cells were provided by Dr. Xiuxing Wang (Nanjing Medical University, Nanjing, China). GSCs were cultured in a neurobasal medium (Gibco, #21103049) supplemented with sodium pyruvate (Gibco, #11360070), GlutaMAX (Gibco, #35050061), B27 minus vitamin A (Gibco, #12587010), 20 ng/mL FGF (R&D Systems, #4114-TC-01M), and 20 ng/mL EGF (R&D Systems, #236-EG-01M). NSCs were cultured in a Neurobasal-A medium without phenol red (Gibco, #12349015) adding the same supplements as added to GSCs. Fluorescently labeled U251 cells (GFP+ U251) were purchased from Shanghai Meixuan Biotechnology (#MXBC260). TAMs isolated from GBM tissues and U251 cells were cultured in a DMEM supplemented with 10% FBS (Gibco, #10099141C). Monocytes and T cells purified from the peripheral blood of healthy individuals and THP1 cells were cultured in an RPMI-1640 medium with L-glutamine (Gibco, #C11875500BT) supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin (Gibco, #15140122). All cells were cultured in a humidified incubator at 37°C with 5% CO2.
Isolation of TAMs from human GBM tumor tissues
Fresh sterile tumor specimens were obtained from patients with GBM who underwent tumor resection surgery. Tissues were washed with 5 mL phosphate-buffered saline (PBS; Gibco, #10010023), minced using scalpels, and digested using a Tumor Dissociation Kit (Miltenyi Biotec, #130-095-929) at 37°C for 30 minutes. Afterward, the sample was filtered through a 70-μm cell strainer (SORFA, #251200) and centrifuged at 300 × g for 5 minutes. Red blood cells were removed using a red blood cell lysis solution (Miltenyi Biotec, #130-094-183). Large amounts of cell debris were removed by a debris removal solution (Miltenyi Biotec, #130-109-398). Subsequently, TAMs were positively selected by CD11b microbeads (Miltenyi Biotec, #130-049-601) following the manufacturer's instructions.
Isolation of monocytes and T cells from peripheral blood
Peripheral blood was collected from healthy volunteers in blood collection tubes containing EDTA (BD Vacutainer, #367525). Peripheral blood mononuclear cells were obtained by Ficoll–Hypaque density gradient centrifugation (GE Healthcare, #17-1440-02) and resuspended in 80 μL autoMACS running buffer (Miltenyi Biotec, #130-091-221). Then, 20 μL CD3 microbeads (Miltenyi Biotec, #130-097-043) were added to positively select CD3+ T cells using an LS column (Miltenyi Biotec, 130-042-401). Monocytes were isolated from CD3– cells using CD14 microbeads (Miltenyi Biotec, #130-050-201) according to the manufacturer's instructions.
Differentiation from monocyte to macrophages
Monocytes were isolated from peripheral blood, then 50 ng/mL macrophage colony-stimulating factor (M-CSF; PeproTech, #300-25) was added into the culture medium, and M0 macrophages were harvested after 6 days.
To obtain M1 macrophages from THP1 cells, THP1 cells were treated with 5 ng/mL phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich, #P8139) for 24 hours. Subsequently, THP1 cells were cultured in a PMA-free media for 72 hours, and then supplemented with 20 ng/mL IFNγ (Novoprotein, #C014) and 250 ng/mL LPS for 48 hours to differentiate into M1 macrophages.
Determination of double-positive TAMs by FACS
CD11b+ cells were isolated from GBM tissues as described above and cultured in a 6-well plate (Corning, #3516) for 3 days (1 × 105 cells/well). GFP+ U251 cells were digested with trypsin and placed at room temperature overnight to prevent adherence. The adherence-depleted U251 cells were washed three times with PBS and added to coculture with macrophages for 24 hours at 37°C. At the end of coculture, adherence-free U251 cells were removed and macrophages were digested from the dish surface and blocked with 1 mL PBS containing 3% BSA at 4°C for 20 minutes. Cells were incubated with the following primary antibodies at 4°C for 1 hour: TMEM119 (clone A16075D; BioLegend, #853301, RRID:AB_2734646, 1:200) and CD49d (clone 9F10; BD Biosciences, #555503, RRID:AB_395893, 1:200). The cells were incubated with the secondary antibody donkey anti-mouse IgG H&L (Alexa Fluor 647; Abcam, #ab150107, RRID:AB_2890037, 1:400) at 4°C for 2 hours. The cells were resuspended in 300 μL PBS and analyzed on a BD FACSVerse Flow Cytometer. Data were acquired using the FlowJo software (version X 10.07r2, RRID:SCR_008520). Double-positive TAMs were determined as GFP+ cells within macrophages, that is, CD49d+GFP+ (phagocytosis by BMDMs) and TMEM119+GFP+ (phagocytosis by MG) double-positive cells.
1 × 106 THP1-derived M1 macrophages were added to coculture with 2 × 106 GFP+ U251 for 2 hours. Then, cells were stained with Brilliant Violet 605 anti-human CD45 (BioLegend, #304042, RRID:AB_2562106). The cells were resuspended in PBS and analyzed on a Flow Cytometer. Double-positive cells were determined as GFP+ cells within the CD45+ cell population.
Phagocytosis inhibition
The phagocytic ability of BMDMs was inhibited by incubation with 1 μmol/L cytochalasin D (Invitrogen, #PHZ1063) for 1 hour at 37°C.
Activation and expansion of T cells
Activated T cells were obtained as previously described (9). Briefly, purified CD3+ T cells were stained with CellTrace Violet Cell Proliferation Kit (Invitrogen, #C34557), cultured in a medium supplemented with 10 ng/mL recombinant human IL2 (PeproTech, #200-02), and activated by coating with 1 μg/mL Ultra-LEAF purified anti-human CD3 antibody (BioLegend, #317325, RRID:AB_11147370) and 5 μg/mL CD28 antibody (BioLegend, #302933, RRID:AB_11150591) for 3 days.
Monocytes were isolated from peripheral blood and differentiated into macrophages. Then, the macrophages were separated into five groups and treated as follows: (i) 1 × 105 macrophages cultured in RPMI-1640 medium with 10% FBS; (ii) 1 × 105 macrophages were treated with 1 μmol/L cytochalasin D for 1 hour at 37°C; (iii) 1 × 105 macrophages noncontact cocultured with 2 × 105 U251 by Transwell with 0.4-μm pore (Corning, #3413) for 24 hours, with U251 placed in the upper chamber, and BMDMs placed in the lower chamber; (iv) 1 × 105 macrophages pretreated with 1 μmol/L cytochalasin D for 1 hour at 37°C and then cocultured with 2 × 105 U251 for 24 hours; (v) 1 × 105 macrophages cocultured with 2 × 105 U251 for 24 hours.
Macrophages that underwent the above treatments were then cocultured with CellTrace-labeled T cells at 37°C in an RPMI-1640 medium supplemented with 10% FBS and 1% penicillin/streptomycin for 3 days. Subsequently, free-floating T cells were collected and analyzed using flow cytometry. Proliferative T cells were identified using the CellTrace signal. The immunosuppressive activity of macrophages was measured by the percentage of proliferative T cells among all T cells. The higher the percentage is, the lower is the immunosuppressive activity of macrophages.
ELISA
THP1-derived M1 macrophages were treated as follows: (i) 1 × 106 macrophages cultured alone; vi) 1 × 106 macrophages cocultured with 2 × 106 NSCs for 24 hours; (iii) 1 × 106 macrophages cocultured with 2 × 106 GSCs for 24 hours; (iv) 1 × 106 macrophages noncontact cocultured with 2 × 106 GSCs for 24 hours by 0.4 μmol/L Transwell, with GSCs placed in the upper chamber and BMDMs placed in the lower chamber; (v) 1 × 106 macrophages pretreated with 1 μmol/L cytochalasin D for 1 hour at 37°C and then cocultured with 2 × 106 U251 for 24 hours. ELISA and flow cytometry were performed on these cells.
1 × 106 THP1-derived M1 macrophages were cocultured with 2 × 106 GFP+ U251 for 24 hours, and then cells were stained with Pacific Blue anti-human CD45 (BioLegend, #982306, RRID:AB_2650649). CD45+GFP+ and CD45+GFP– cells were sorted by fluorescence-activated cell sorting (FACS) for ELISA and flow cytometry analysis.
The protein levels of IL1α, IL1β, TNFα, and IL10 in the macrophage culture supernatant were measured with Human IL1α ELISA Kit (Cusabio, #CSB-E04620h), Human IL1β ELISA Kit (Cusabio, #CSB-E08053h), Human TNFα ELISA Kit (Cusabio, #CSB-E04740h), and Human IL10 ELISA Kit (Cusabio, #CSB-E04593h), respectively. Experiments were conducted according to the manufacturer's instructions.
Flow cytometry
Cells were stained with APC anti-human CD163 Antibody (BioLegend, #333610, RRID:AB_2074533), Alexa Fluor 647 anti-human CD206 (MMR) Antibody (BioLegend, #321116), PE anti-human CD274 (B7-H1, PD-L1) Antibody (BioLegend, #329706, RRID:AB_940368), APC anti-human CD273 (B7-DC, PD-L2) Antibody (BioLegend, #329708), PE anti-human CD276 (B7-H3) Antibody (BioLegend, #351004, RRID:AB_10720987), and Pacific Blue anti-human CD45 (BioLegend, #982306, RRID:AB_2650649) following the manufacturer's instructions.
Locate the phagocytosed tumor DNA in macrophages
A total of 7 × 105 THP1-derived M1 macrophages were treated with 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO; Beyotime, #C1993S) at 37°C for 15 minutes to stain the plasma membrane. 1.4 × 106 U251 were incubated at 55°C water for 20 minutes and stained with propidium iodide (PI) for 5 minutes at room temperature. M1 macrophages and U251 were cocultured under the serum-free medium for 2 hours. Cells were fixed with 4% paraformaldehyde (PFA) for 10 minutes at room temperature and treated with Antifade Mounting Medium with DAPI (Beyotime, #P0131). Images were captured with Nikon A1R confocal microscopy.
Live-cell imaging system
5 × 104 GFP+ U251 cells were cocultured with 2.5 × 104 THP1-derived M1 macrophages for 2 hours. Time series images were captured by Opera Phenix Plus High-Content Screening System every 3 minutes and a total of 200 time points were obtained for each view. Time-series images were processed into a video by Harmony software (version 4.9).
Bulk RNA isolation and sequencing
1 × 106 GSCs/NSCs were digested with accutase (BioLegend, #423201) and placed at room temperature overnight to prevent adherence. GSCs/NSCs were washed with PBS three times and added to the dish of macrophage for coculture. BMDMs were separated into four groups and treated as follows: (i) 1 × 106 BMDMs cultured alone; (ii) 1 × 106 BMDMs cocultured with 2 × 106 adherence-depleted NSCs for 24 hours; (iii)1 × 106 BMDMs cocultured with 2 × 106 adherence-depleted GSCs for 24 hours; (iv) 1 × 106 BMDMs were treated with 1 μmol/L cytochalasin D for 1 hour at 37°C, then cocultured with 2 × 106 adherence-depleted GSCs for 24 hours. At the end of coculture, adherence-free cells were removed, and BMDMs that underwent the above treatments were harvested for RNA-seq.
Total RNA was extracted from BMDMs using TRIzol Reagent (Invitrogen, #15596026) according to the manufacturer's instructions. The quality of the libraries was measured using an Agilent 2100 Bioanalyzer. The length and distribution of fragments were verified via an Agilent High Sensitivity DNA kit (Agilent, #5067-4626). qPCR was performed on a Thermo Scientific StepOnePlus Real-Time PCR System to determine the effective concentration of the libraries. Finally, multiplexed paired-end reads were sequenced on the Illumina NovaSeq 6000 platform.
Smart-seq2 library construction for mouse double-positive cells
All mouse experiments were performed under the guidelines of and were approved by the Animal Care and Use Committee of Nanjing Medical University. C57BL/6J mice (RRID:IMSR_JAX:000664; 7 weeks of age) were purchased from The Jackson Laboratory. Bone marrow cells were isolated from the femurs and tibias of C57BL/6J mice, and filtered through a 70-μm cell strainer, then centrifuged at 1,500 rpm for 3 minutes, and the supernatant was removed. Red blood cells were removed by red blood cell lysis solution, then centrifuged at 1,500 rpm for 3 minutes, and the supernatant was removed. Cells were resuspended with DMEM supplemented with 20 ng/mL M-CSF (PeproTech, #315-02) and cultured for 7 days. On the 7th day, cells were cultured with a fresh stimulation medium (DMEM containing 10% FBS and 100 ng/mL LPS) for 24 hours to obtain M1 macrophages.
1 × 106 M1 macrophages were cocultured with 2 × 106 GFP+ U251 for 2 hours. Cells were stained by F4/80 (BioLegend, #123110, RRID:AB_893486) and sorted with FACS. 1 × 103 F4/80+GFP+ cells and 1 × 103 F4/80+GFP– cells were collected for Smart-seq2 library construction and performed RNA-seq on the Illumina NovaSeq 6000 platform.
scRNA-seq data analysis (Fluidigm C1 platform)
Raw reads were trimmed using Trimmomatic (version 0.38, RRID:SCR_011848; ref. 13) to obtain clean reads. Subsequently, reads were mapped to the GRCh37 human genome using HISAT2 (version 2.1.0; ref. 14) with default parameters, and gene expression was quantified using featureCounts (15) in the Subread package (version 1.6.3) with parameters –p –t exon –g gene_id. Low-quality cells were removed if the number of detected genes was less than 800 or the number of aligned reads was less than 50,000. Finally, we retained 346 cells, of which, expression levels were measured as counts per million (CPM). Dimensionality reduction and clustering were performed using the R package Monocle2 (version 2.12.0; ref. 16).
For mutation analysis, clean reads were mapped to the GRCh37 human genome using STAR (version 2.6.1b; ref. 17) in two-pass mode with the parameter –outFilterScoreMinOverLread 0.1 –outFilterMatchNminOverLread 0.1. Then, we marked the duplicates using Picard (https://github.com/broadinstitute/picard) MarkDuplicates (version 2.18.11), followed by GATK (version 4.0.7.0, RRID:SCR_001876; ref. 18) tools SplitNCigarReads, BaseRecalibrator and HaplotypeCaller. Raw variants were filtered by GATK tools VariantFiltration with the following parameters: -window 35 –cluster 3 –filterName FS –filter “FS > 30.0” –filterName QD –filter “QD < 2.0.” A tumor-derived mutation at the single-cell level was defined by the following criteria: Mutations called by scRNA-seq were maintained if these mutations were also detected by WES and each mutation must recurrently appear in more than three cells.
Copy-number variations (CNV) inferred from scRNA-seq were called using the R package infercnv (19) with the parameters denoise = TRUE and HMM = TRUE. The heat maps displaying the CNVs of the cells were created using the ComplexHeatmap (version 2.0.0; ref. 20). The initial CNV matrix was obtained using infercnv, with rows representing genes, and columns representing cells. Values in the matrix were restandardized to –1 and 1, and the CNV levels of each cell were calculated as the quadratic sum of all genomic regions.
scRNA-seq data analysis (10X Genomics platform)
The raw gene-expression matrix was generated using the CellRanger pipeline (version 3.0.2). Then, the matrices of all samples were combined with R (version 3.6.0) and converted to the Seurat object using the R package Seurat (version 3.2.2; ref. 21). Cells isolated from different patients were integrated using the R package rliger (version 1.0.0; ref. 22). Cells were filtered out if the expression of mitochondrial genes was greater than 10% or if detected genes were less than 800 or greater than 7,500. Finally, we retained 17,144 cells for the downstream analysis, of which, 8,270 cells were identified as tumor cells based on the expression of SOX9 and BCAN. 876 cells were identified as oligodendrocytes based on the expression of MOG and MAG, 74 cells were identified as endothelial cells based on the expression of MCAM and ESAM, and 137 cells were identified as T cells based on the expression of PTPRC and CD3D. Macrophages were further annotated as BMDMs (LYZ and TGFBI, n = 3,535) and MG (P2RY12 and TMEM119, n = 4,252).
WES data analysis
Clean DNA reads were mapped to the GRCh37 human genome using BWA (version 0.7.15, RRID:SCR_010910; ref. 23), and the sorted bam files were deduplicated using Picard tools. The GATK tools RealignerTargetCreator, IndelRealigner, BaseRecalibrator, and Mutect2 were applied successively to call somatic mutations. The raw VCF files were filtered using the GATK tools CalculateContamination and FilterMutectCalls.
CNVs were called using the Python package CNVkit (version 0.9.7.b1; ref. 24) with parameters –drop-low-coverage –scatter –diagram –method amplicon.
Analysis of CyTOF data
Clinical information of GBM patients involved in this study was listed in Supplementary Table S1. The CyTOF data were processed using the R package cytofWorkflow (version 1.14.0; ref. 25). FCS files of GBM patients were loaded into R using the read.flowSet function. A total of 20,000 cells were randomly selected if more than 20,000 cells were detected in the patient; otherwise, all cells were kept for further analysis.
Single cells were annotated based on the following markers: B cells (CD19+CD3–CD11b–), plasma cells (CD3–CD19–CD38high), T cells (CD3+), macrophages (CD64+CD3–CD19–), dendritic cells (DC, CD11b+CD1c+), neutrophils (CD66b+CD11b+), and natural killer (NK) cells (CD56+CD3–CD19–CD11b–). Macrophages/monocytes were further annotated using the following markers: monocyte (CD14+CCR2+), BMDM (CD49d+CX3CR1low), and microglia (CD49d–CX3CR1high). Patients were classified into the MERTK-high and MERTK-low groups according to their expression level of MERTK in TAMs.
Bulk RNA-seq data analysis
For RNA-seq of human macrophages, paired-end reads were trimmed using Trimmomatic and then mapped to the GRCh37 human genome using HISAT2. Quantification of genes was conducted using featureCounts. Differentially expressed genes were calculated using the R package DESeq2 (version 1.24.0, RRID:SCR_000154; ref. 26).
For RNA-seq of mouse macrophages, raw reads were trimmed and then mapped to the mm10 mouse genome using STAR. Quantification of genes was conducted using featureCounts. Differentially expressed genes were calculated using the R package DESeq2.
Calculation of the signature score
The normalized BMDMs, MG, M1/M2, and phagocytosis signature scores were calculated using single-sample gene set enrichment analysis (ssGSEA; ref. 27). All the signatures involved in ssGSEA are listed in Supplementary Table S2.
GSEA
GSEA was conducted using the GSEA function in the R package clusterProfiler (version 3.16.1; ref. 28). The input genes were ranked according to log2FoldChange as determined using DESeq2. P values were adjusted using the BH method.
Pathway enrichment analysis
Pathway enrichment analysis was conducted using enrichGO tools in the R package clusterProfiler, with parameters ont = “BP” and pAdjustMethod = “BH.”
Survival analysis
Patients were classified into high and low groups according to the median of the normalized enrichment score of the unified BMDMs/MG signature. Kaplan–Meier survival curves were generated using the R package survminer (version 0.4.8) to estimate the overall survival difference between groups, and P-values were calculated using the log-rank test.
Statistical analysis
Detailed statistical information is indicated in the corresponding methods or figure legends. Asterisks are used to indicate statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001); n.s., statistically nonsignificant (P > 0.05).
Data and codes availability
Raw RNA-seq data generated in this study are available from Sequence Read Archive via accession number PRJNA911481. The WES data analyzed in this study were obtained from European Genome-phenome Archive (EGA) with accession number EGAS00001001900 (8). The scRNA-seq data of Fluidigm C1 were obtained from EGA with accession number EGAS00001002185 (8). The scRNA-seq data of 10X Genomics were provided by Couturier and colleagues (29) and were available from https://github.com/mbourgey/scRNA_GBM. The CyTOF data analyzed in this study were provided by Friebel and colleagues (30) and were available from https://data.mendeley.com/datasets/jk8c3c3nmz/draft?a=c0a9d8dc-8ac2–4942-baf9–208de7a8c310. Computational codes used for this study are accessible at GitHub: https://github.com/biowumin/Double-positive-TAMs.
Results
Identification of double-positive cells with macrophage–tumor signature
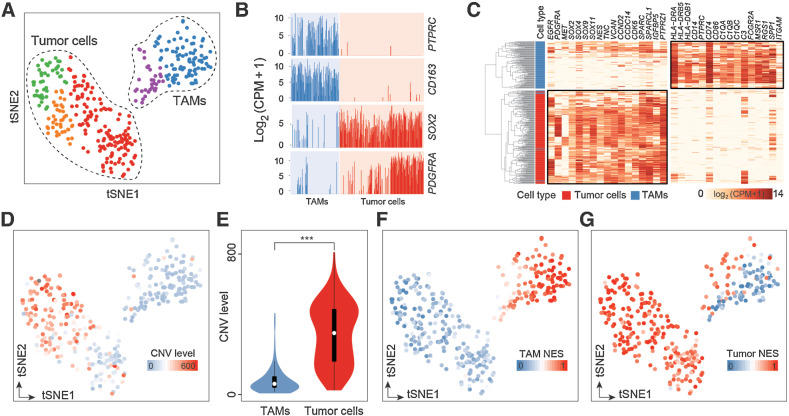
We integrated scRNA-seq data and matched WES data of three GBM patients from a published study (8), which included 384 tumor cells and CD11b-purified TAMs (Supplementary Table S1). After quality control, a total of 346 single cells were retained for the subsequent analysis. To create a comprehensive view of the major population structure, we generated a two-dimensional map using t-distributed stochastic neighbor embedding (t-SNE) and classified cells into five major clusters by unsupervised clustering (Fig. 1A). We did not observe significant interpatient batch effects (Supplementary Fig. S1A–S1B).
Figure 1.
scRNA-seq analysis identifies a TAM subset that dually expresses tumor and macrophage signatures. A, t-SNE map showing the clustering of 346 cells. Each dot represents one cell colored by clusters. B, Bar plots displaying the mRNA expression levels of the indicated markers, measured by log2-transformed CPM. Each bar represents a single cell, colored by cell types identified in A. C, Hierarchical clustering of 346 cells (rows) according to the cell type–specific markers (columns). D, t-SNE map showing the CNV levels of 346 cells. Cells are colored by CNV levels as determined by R package infercnv. E, Violin plots showing the CNV levels of TAMs (n = 125) and tumor cells (n = 221). P values are calculated by a two-sided Wilcoxon rank-sum test. F and G, t-SNE expression map, colored by the NES of TAM-specific gene signature (F) and tumor-specific gene signature (G). NES was calculated using ssGSEA. ***, P < 0.001.
We annotated the cell clusters according to the canonical markers. Clusters that expressed PTPRZ1, NES, EGFR, PDGFRA, and SOX2 were identified as tumor cells (n = 221), whereas clusters that expressed HLA-DRA, FCGR1A, CD14, CD163, and PTPRC were identified as macrophages (n = 125; Fig. 1B; Supplementary Figs. S1C-S1E). Hierarchical clustering of these cells according to literature-based gene signatures (8) also showed that the identified cell populations had different transcriptomic features. Tumor cells were characterized by the expression of glioma cell markers (Supplementary Table S2). In contrast, TAMs were characterized by the expression of immune cell markers PTPRC, macrophage markers (CD14, ITGAM, FCGR2A, and MSR1/CD204), MHC class II molecules (HLA-DRA, HLA-DRB5, and HLA-DQB1), and complement components (C1QA, C1QB, C1QC, and C3; Fig. 1C).
To further confirm the identities of the identified cell populations, we inferred chromosomal CNVs based on scRNA-seq data. Tumor cells harbored the amplification of chromosome 7 and deletion of chromosome 10, both of which are the prominent genomic characteristics of GBM. These results were concordant with matched WES results (Supplementary Fig. S1F). Tumor cells showed significantly higher CNV levels than TAMs (median CNV level: 349.76 vs. 60.84, P < 2.2e−16; Fig. 1D and E). GSEA using MSigDB Hallmark gene sets (31) revealed that tumor cells were enriched in the following metabolic processes (adipogenesis: normalized enrichment score (NES) = 2.14, q = 0.0008; fatty acid metabolism: NES = 2.23, q < 0.0001; oxidative phosphorylation: NES = 2.22, q < 0.0001; glycolysis: NES = 2.09, q = 0.0007), cell proliferation (mitotic spindle: NES = 2.07, q = 0.0009), and DNA repair (NES = 2.14, q = 0.0008; Supplementary Fig. S1G). TAMs were enriched in immune-associated pathways, such as TNFα signaling via NFκB (NES = −2.06, q = 0.0002, IFNγ response (NES = −1.61, q = 0.028), inflammatory response (NES = −2.43, q < 0.0001), and complement (NES = −1.59, q = 0.02; Supplementary Fig. S1H).
Interestingly, we identified a subset of cells that expressed both macrophage and tumor signatures simultaneously (Fig. 1F and G). High expression of a macrophage signature (TAM NES > 0.5) was observed in 96% (120/125) of the TAM population, whereas it was observed in only 0.45% (1/221) of the tumor population (Fig. 1F). High expression of a tumor signature (tumor NES > 0.5) was observed in 99.55% (220/221) of the tumor population. Notably, 35.2% (44/125) of TAMs also highly expressed a tumor signature (Fig. 1G). We defined TAMs with both tumor NES and TAM NES greater than 0.5 as double-positive TAMs (Supplementary Fig. S2A). In total, we identified 39 double-positive TAMs.
Double-positive TAMs exhibit unique transcriptome characteristics
To further characterize the double-positive TAMs, we integrated scRNA-seq data with matched WES data and found that double-positive TAMs harbored GBM-derived mutations (see Materials and Methods). WES data of patient SF10282 showed that a somatic mutation (chr4:55133837 G>A) was located in PDGFRA (Fig. 2A). Meanwhile, the scRNA-seq data of patient SF10282 showed that cells SF10282-E1 and SF10282-G3, identified as TAMs, also included the mutation chr4:55133837 G>A (Fig. 2A). This was unlikely due to cell misclassification because the canonical macrophage markers (CD163, FCGR2A, MSR1, and ITGAX) were highly expressed in both SF10282-E1 and SF10282-G3, whereas they were rarely expressed in tumor cells (Fig. 2B). We identified a total of 12 TAMs that harbored GBM-derived mutations (Supplementary Table S3 and Supplementary Fig. S2B), and all these TAMs were double-positive TAMs (12/39 in double-positive TAMs, 0/86 in other TAMs). Next, we compared the difference in CNV levels between double-positive TAMs and other TAMs (Fig. 2C). Double-positive TAMs showed significantly higher CNV levels (median CNV level: 112.40 vs. 51.53, P = 2.57e−10; Fig. 2D).
Figure 2.
Characterization of expression profiles in double-positive TAMs. A, Double-positive TAMs harbor a tumor-derived mutation (chr4:55133837 G>A) located in PDGFRA. Top, schematic diagram showing the human reference genome sequence from chr4:55133833–55133841. Middle, WES results of patient SF10282; bars represent sequencing reads from tumor tissues and matched whole blood. Bottom, scRNA-seq results of cells SF10282-E1 and SF10282-G3. Bars are colored according to the mutation status of the 55133837th base (red, mutant; blue, nonmutant). The height of the bars is proportional to the number of reads. Coverage, the number of reads mapped to the 55133837th base; QV, average sequencing quality score of the reads; MAPQ, average mapping score. B, Bar plots showing the mRNA expression levels of the indicated markers. Bars are colored according to annotated cell types: red, tumor cells; blue, TAMs; black, indicated single cell. Data are presented as the mean ± SEM. C, Heat map showing the CNV levels (red, amplification; blue, deletion). Rows represent chromosomal locations and columns represent cells. D, Violin plot showing the CNV levels of double-positive TAMs and other TAMs. E, Heat map depicting the differentially expressed genes between double-positive TAMs and other TAMs. F and G, GO terms enriched by double-positive TAMs (F) and other TAMs (G). Bars are colored by cell types. APP, antigen processing and presentation; CHO, carbohydrate; PS, polysaccharide; Reg., regulation. Overrepresentation analysis was performed, and q values were calculated via the R package clusterProfiler. H and I, Bar plots displaying the mRNA expression of MG- (H) and BMDM-like (I) markers in the TAM subsets. Bars are colored by cell types. Data are presented as the mean ± SEM. P values were calculated by a two-sided Wilcoxon rank-sum test. *, P < 0.05; **, P < 0.01; ***, P < 0.001; n.s., nonsignificant.
We compared the expression pattern between the two TAM populations. Genes upregulated in double-positive TAMs included lysosomal proteases (CTSD and CTSA; ref. 32), T cell–response regulators [ENTPD1/CD39 (33), CD274/PD-L1 (34), and LGALS3 (35)], transforming growth factor-β (TGFB2 and TGFB3; ref. 35), and M2 polarization genes (FN1, MARCO, and LYVE1; Fig. 2E; ref. 36). These genes were enriched in Gene Ontology (GO) terms including negative regulation of macrophage cytokine production, negative regulation of immune effector process, and negative regulation of immune response (Fig. 2F), implying the immunosuppressive phenotype of double-positive TAMs. Genes upregulated in the other TAM population included MHC class II molecules (HLA-DRA and HLA-DRB1), M1 polarization genes (IL1A, IL1B, CCR7, and TNF; ref. 36), costimulatory molecule (CD86), and Fc gamma receptor gene (FCGR1A/CD64). These genes were enriched in GO terms including positive regulation of cytokine biosynthetic process, positive regulation of humoral immune response, IL2 biosynthetic process, positive regulation of acute inflammatory response, and antigen presentation via MHC class II (Fig. 2G), implying the active immune response. Given that TAMs are composed of BMDMs and MG in the GBM microenvironment, we checked the expression levels of canonical BMDM and MG markers to determine the identity of double-positive TAMs. In comparison with MG markers (P2RY13, SCIN, OLFML3, and CX3CR1; Fig. 2H; ref. 37), double-positive TAMs highly expressed well-established BMDM markers (FN1, AHR, TGFBI, and ITGA4; Fig. 2I; ref. 38), implying that double-positive TAMs tended to be BMDM-like. Collectively, we found that double-positive TAMs harbored tumor-derived mutations and exhibited immunosuppressive phenotypes. Ontogenetically, double-positive TAMs tended to be BMDM-like, rather than MG-like.
Validation of double-positive TAMs
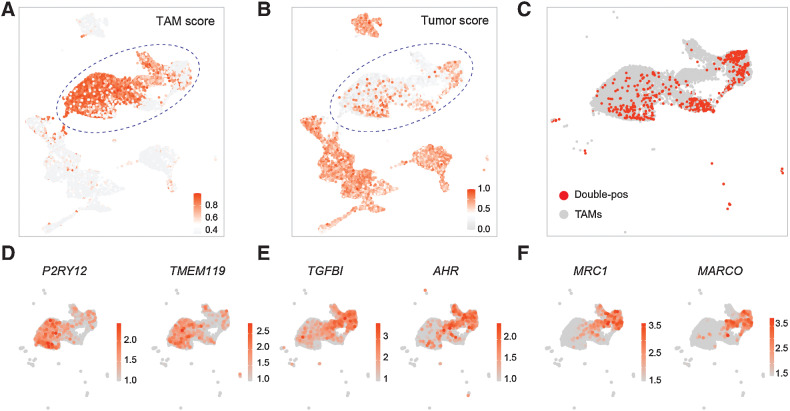
To validate the existence of double-positive TAMs, we used an independent scRNA-seq data set, which included seven IDH wild-type GBM patients (Supplementary Table S1; ref. 29). After integration and quality control, we obtained 17,144 cells clustered into 21 clusters for downstream analysis (Supplementary Fig. S3A–S3B). We annotated these clusters based on canonical cell type–specific markers: 8,270 tumor cells (SOX9 and BCAN), 876 oligodendrocytes (MOG and MAG), 74 endothelial cells (MCAM and ESAM), 137 T cells (PTPRC and CD3D), 4,252 MG (P2RY12 and TMEM119), and 3,535 BMDMs (LYZ and TGFBI; Supplementary Fig. S3C–S3D). High expression of a macrophage signature was observed in 79% (6,176/7,787) of the TAMs population, whereas it was observed in only 9% (736/8,270) of the tumor population (Fig. 3A). High expression of a tumor signature was observed in 88% (7,290/8,270) of the tumor population. Notably, 12% (935/7,787) of TAMs also highly expressed a tumor signature (Fig. 3B). We defined TAMs with tumor scores greater than 0.45 as double-positive TAMs (Supplementary Fig. S3E). A total of 935 double-positive TAMs were identified, of which, 678 (72.51%) were BMDMs and 257 (27.49%) were MG (Fig. 3C–E). Besides, we found the double-positive TAMs highly expressed classic M2 macrophage markers MRC1 and MARCO (Fig. 3F). These data further validated the existence of double-positive TAMs, their BMDM identity, and immunosuppressive phenotypes, which were consistent with our previous observations.
Figure 3.
Validation of double-positive TAMs. A and B, Uniform Manifold Approximation and Projection (UMAP) plot displaying the expression of TAMs (A) and tumor (B) signature score, colored by the normalized signature score level. C, UMAP plot displaying the distribution of double-positive (double-pos) TAMs, colored by cell types. D and E, UMAP plot displaying the expression of canonical MG (D) and BMDMs (E) markers, colored by the expression levels of indicated genes. F, UMAP plot displaying the expression levels of M2 macrophage markers, colored by the expression level of indicated genes.
Generation of double-positive TAMs by coculture BMDMs with glioma cells
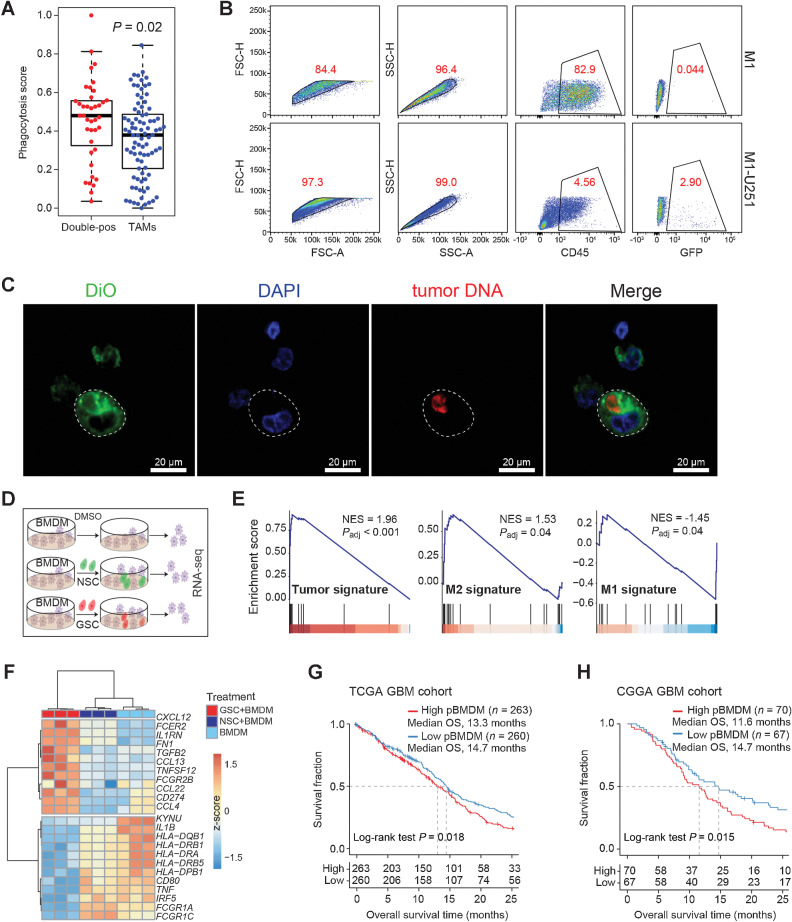
Macrophages are professional phagocytes, therefore, we hypothesized that the existence of double-positive TAMs was associated with phagocytosis of glioma cells. We compared the phagocytic capacity between double-positive TAMs and other TAMs and found that double-positive TAMs showed higher phagocytic features (median phagocytosis score: 0.48 vs. 0.38, P = 0.02; Fig. 4A). We also observed higher expression of lysosomal proteases (CTSD and CTSA), responsible for the degradation of phagocytosed proteins, in double-positive TAMs (Fig. 2E). These results indicated the correlation between phagocytosis and double-positive TAMs. We isolated CD11b+ TAMs from tumor tissues of two newly diagnosed GBM patients (Supplementary Table S1), then cocultured them with GFP+ U251 (green fluorescent protein-labeled U251 cells), and utilized FACS to demonstrate the existence of double-positive TAMs (Supplementary Fig. S4A). We found that 85% of CD49d+TMEM119low BMDMs harbored GFP signals, whereas only 0.21% of CD49d–TMEM119high MG harbored GFP signals (Supplementary Fig. S4B), indicating that glioma cells were more likely to be engulfed by BMDMs than MG.
Figure 4.
Formation of the double-positive TAMs in an in vitro coculture system. A, Box plot showing the phagocytosis score of double-positive TAMs (n = 39) and other TAMs (n = 86). P values are calculated by the two-sided Wilcoxon rank-sum test. B, Flow cytometry analysis showing the GFP fluorescence in THP1-derived M1 macrophages cultured alone or cocultured with GFP+ U251 cells. C, Representative fluorescence images of M1 macrophages. Tumor DNA in U251 was prelabeled by PI, and the plasma membrane of M1 macrophages was labeled with 3,3′-dioctadecyloxacarbocyanine perchlorate (DiO). Subsequently, the U251 cells were cocultured with M1 macrophages for 2 hours, and then DNA of M1 macrophages was labeled by 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 20 μm. D, Schematic workflow for the RNA-seq of BMDMs. BMDMs were cocultured with DMSO (top), NSCs (middle), or GSCs (bottom) for 24 hours, and total RNA was extracted for sequencing. E, GSEA plots of BMDMs cocultured with GSCs compared with BMDMs cocultured with NSCs. GSEA was performed by R package clusterProfiler. F, Heat map depicting the differentially expressed genes following different treatments. G and H, Kaplan–Meier survival curves show the OS of GBM patients in the TCGA (G) and CGGA cohorts (H). Patients are classified into two groups based on the median NES of the pBMDMs signature. P values were calculated by the log-rank test.
To generate double-positive TAMs in vitro, we cocultured THP1-derived macrophages with GFP+ U251 cells for 2 hours and stained cells with CD45 antibody. We checked the double-positive TAMs by FACS. There were 2.9% CD45+ BMDMs harboring GFP fluorescence, indicating the macrophages phagocytosed U251 cells (Fig. 4B). We also confirmed the existence of double-positive TAMs by a live-cell imaging system. As shown in the Supplementary Video, we found that tumor cells were engulfed by macrophages (green fluorescence within macrophages).
To exclude the possibility that the double-positive TAMs were formed by cell fusion, where the nucleus of the target cell fuses with the nucleus of host cells (39), we performed confocal analysis and found that the engulfed tumor DNA (red) located in the cytoplasm (green) of macrophages, and there was no colocalization of the tumor DNA (red) and the macrophage DNA (blue), indicating that the nucleus of U251 cells did not fuse with the nucleus of macrophages (Fig. 4C). These data showed that the double-positive TAMs were not formed by cell fusion.
Double-positive TAMs exhibit immunosuppressive phenotype
To explore the immune phenotype of double-positive TAMs generated in vitro, we analyzed the mRNA expression profiles of BMDMs cocultured with GSCs/NSCs or cultured alone (Fig. 4D). Compared with the BMDMs cultured alone, the M2 signature (NES = 1.58, P = 0.026) and tumor signature (NES = 2.11, P < 0.001) were significantly upregulated, whereas the M1 signature (NES = –1.72, P = 0.0064) was significantly downregulated in BMDMs cocultured with GSCs (Supplementary Fig. S4C). We observed no significant difference in M2 and tumor signatures between BMDMs cocultured with NSCs and BMDMs cultured alone (Supplementary Fig. S4D). When comparing with BMDMs cocultured with NSCs, the significant increase in tumor signature (NES = 1.96, P < 0.001) and M2 signature (NES = 1.53, P = 0.04) accompanied by the decrease of the M1 signature (NES = –1.45, P = 0.04) was observed in BMDMs cocultured with GSCs (Fig. 4E). Chemokine genes (CXCL12, CCL13, CCL22, and CCL4), Fc receptors (FCER2/CD23 and FCGR2B/CD32), cytokines (TNFSF12 and IL1RN), and immune inhibitory receptor–ligand CD274/PD-L1 were upregulated in BMDMs cocultured with GSCs compared with that in BMDMs cocultured with NSCs or cultured alone (Fig. 4F). Functional enrichment analysis showed that these genes were enriched in biological processes such as negative regulation of cell–cell adhesion, leukocyte migration, and negative regulation of cytokine production (Supplementary Fig. S4E). However, MHC class II molecules (HLA-DRB5, HLA-DRA, HLA-DRB1, HLA-DPB1, and HLA-DQB1), Fc gamma receptor genes (FCGR1A and FCGR1C), and M1 polarization genes (TNF, IL1B, KYNU, CD80, and IRF5) were downregulated in BMDMs cocultured with GSCs compared with that in BMDMs cocultured with NSCs or cultured alone (Fig. 4F). These genes were enriched in biological processes such as response to interferon-gamma, antigen process and presentation, and the T-cell receptor signaling pathway (Supplementary Fig. S4F). The above findings collectively verified the immunosuppressive activity of double-positive TAMs.
We defined a unified BMDM (pBMDM) signature as the combination of BMDMs and phagocytosis signature and correlated pBMDM signature with the overall survival (OS) of GBM patients. We used the publicly available The Cancer Genome Atlas (TCGA)-GBM cohort (40) and observed a significantly longer OS (median OS: 14.7 vs. 13.3 months; P = 0.018) in patients with a lower pBMDM signature than those with higher pBMDM signature (Fig. 4G). The same findings were observed in the Chinese Glioma Genome Atlas (CGGA) cohort (median OS: 14.7 vs. 11.6 months; P = 0.015; Fig. 4H; ref. 41).
Immunosuppressive phenotype of double-positive TAMs is caused by phagocytosis of glioma cells
To investigate whether the immunosuppressive activity of double-positive TAMs was caused by phagocytosis of glioma cells, we treated BMDMs with cytochalasin D (cytoD), an actin polymerization inhibitor that can inhibit the phagocytic ability of BMDMs. We compared the mRNA expression profiles of cocultured BMDMs with cytoD treatment or not (Fig. 5A). M2 signature (NES = −1.66, P = 0.01) and tumor signature (NES = −1.98, P < 0.001) were significantly downregulated in cytoD-treated BMDMs, whereas the M1 signature showed no significant difference (NES = 1.97, P = 0.23; Fig. 5B). The expression levels of proinflammatory genes IL1B, TNF, IL18, and IL23A were significantly downregulated (Student t test, P < 0.05) in BMDMs cocultured with GSCs compared with that in BMDMs cultured alone. The expression levels of these genes had different degrees of recovery in phagocytosis-inhibited (i.e., cytoD-treated) BMDMs (Fig. 5C).
Figure 5.
Immunosuppressive phenotype of double-positive TAMs is caused by phagocytosis of glioma cells. A, Schematic workflow for the RNA-seq of BMDMs. Top, BMDMs cocultured with GSCs for 24 hours. Bottom, BMDMs were pretreated with cytoD for 1 hour to inhibit their phagocytic abilities, then cocultured with GSCs for 24 hours. B, GSEA plots of BMDMs treated with cytoD before coculture compared with BMDMs cocultured with GSCs directly. GSEA was performed by the R package clusterProfiler. C, Bar plots showing the expression levels of proinflammatory genes. Bars are colored according to treatments. Data, mean ± SEM. P values were calculated by two-sided Student t test. D, Heat map depicting the differentially expressed genes following different treatments. E, Comparison of IL1α, IL1β, TNFα, and IL10 levels in M1 BMDMs under different coculture conditions. M1, M1 macrophages cultured alone; M1 + NSC, M1 macrophages cocultured with NSCs; M1 + GSC, M1 macrophages cocultured with GSCs; M1 + GSC#, M1 macrophages cocultured with GSCs by Transwell with 0.4 μm pore; cytoD-M1 + GSC, M1 macrophages pretreated with cytoD, then cocultured with GSCs. Data, mean + SEM. P values were calculated by two-sided Student t test. F, Comparison of mean fluorescence intensity (MFI) of CD163, CD206, CD276, PD-L1, and PD-L2 in M1 BMDMs under different coculture conditions. M1, M1 macrophages cultured alone; M1 + NSC, M1 macrophages cocultured with NSCs; M1 + GSC, M1 macrophages cocultured with GSCs; M1 + GSC#, M1 macrophages cocultured with GSCs by Transwell with 0.4 μm pore; cytoD-M1 + GSC, M1 macrophages pretreated with cytoD, then cocultured with GSCs. Data, mean + SEM. P values were calculated by two-sided Student t test. G, Percentage of proliferative T cells under different treatments. #, cells cultured by Transwell with 0.4 μm pore. Data, mean ± SEM. P values are calculated by a two-sided Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
We checked the expression of activating and inhibitory immune regulatory factors (36) and observed the increased expression of T-cell inhibition mediators (CD274/PD-L1 and PDCD1LG2/PD-L2) and decreased expression of T cell–activating signals (TNFSF4/OX-40L, TNFSF15, CD80, ICOSLG/B7H2, CD40, and CD86) in BMDMs cocultured with GSCs compared with that in BMDMs cultured alone (Fig. 5D). In cytoD-treated BMDMs, the expression levels of T-cell suppressors (CD274/PD-L1, CD276/B7H3, LGALS9, and PDCD1LG2/PD-L2) were decreased, whereas T-cell activators (TNFSF4/OX-40L, CD80, TNFSF15, and ICOSLG/B7H2) were increased, as compared with that in BMDMs directly cocultured with GSCs (Fig. 5D).
Furthermore, we used ELISA to analyze the levels of proinflammatory cytokines (IL1α, IL1β, and TNFα; ref. 42) and anti-inflammatory cytokine IL10 (43) in BMDMs under different coculture conditions, including (i) BMDMs cultured alone, (ii) BMDMs noncontact cocultured with GSCs by Transwell chambers (so that BMDMs cannot phagocytose tumor cells), (iii) cytoD-treated BMDMs cocultured with GSCs, (iv) BMDMs cocultured with NSCs, and (v) BMDMs cocultured with GSCs. The levels of proinflammation cytokines in BMDMs cocultured with GSCs were significantly decreased (Student t test, P < 0.01) compared with other groups (Fig. 5E). When inhibiting the phagocytic ability of BMDMs, then coculturing them with GSCs, the levels of proinflammation cytokines (IL1α, IL1β, and TNFα) and anti-inflammatory cytokine IL10 in these BMDMs tended to be higher and lower (Student t test, P < 0.01), respectively (Fig. 5E). We also measured the levels of M2 macrophage markers (CD163 and CD206; ref. 44) and immune-checkpoint proteins (CD276, PD-L1, and PD-L2; ref. 45) by FACS. BMDMs cocultured with GSCs showed significantly higher expression levels of these proteins than other groups (Student t test, P < 0.05; Fig. 5F). When inhibiting the phagocytic ability of BMDMs that cocultured with GSCs, these proteins significantly decreased (Student t test, P < 0.05; Fig. 5F). These results collectively suggested that the immunosuppressive phenotype of BMDMs was caused by engulfing glioma cells.
To elucidate the immunosuppressive effect of double-positive TAMs on T cells, we cocultured BMDMs with T cells and measured the immunosuppressive ability by the percentage of proliferative T cells (see Materials and Methods). If BMDMs had a suppressive effect on T cells, the percentage of proliferative T cells would decrease. We obtained BMDMs with the following treatments: BMDMs cultured alone (A), BMDMs treated with cytoD (B), BMDMs noncontact cocultured with U251 by Transwell chambers (C), cytoD-treated BMDMs cocultured with U251 (D), and BMDMs cocultured with U251 (E; Supplementary Fig. S5). Next, BMDMs under different treatments were cocultured with activated T cells to measure their immunosuppressive activity (Supplementary Fig. S5). CytoD treatment alone did not affect the proliferation of T cells (A: 82.47% ± 1.66%, B: 75.97% ± 3.44%, P = 0.19), BMDMs cocultured with U251 by Transwell had a suppressive effect on the proliferative T cells (C: 60.83% ± 2.71%, C vs. A P = 0.0047), and BMDMs directly cocultured with U251 significantly reduced the proliferation of T cells (E: 24.5% ± 3.5%, E vs. A P = 0.00084). However, BMDMs pretreated with cytoD before coculturing with U251 had weakened immunosuppressive activity relative to BMDMs that were cocultured with U251 directly (D: 49.27% ± 3.45%, D vs. E P = 0.0073; Fig. 5G). These findings again verified that the immune-suppressive phenotype of double-positive TAMs was caused by the phagocytosis of glioma cells.
Double-positive TAMs drive immunosuppression by immune-checkpoint proteins
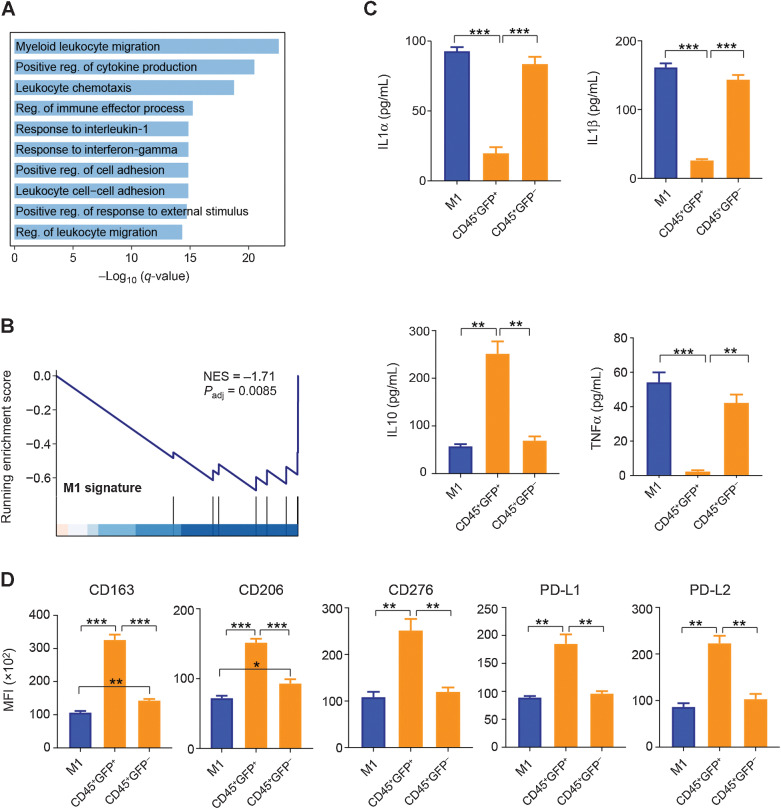
To investigate the immunosuppressive mechanism of double-positive TAMs, we isolated monocytes from mice and differentiated monocytes into M1 macrophages, which were cocultured with GFP+ U251. M1 macrophages were labeled by the F4/80 antibody, and then GFP+F4/80+ and GFP–F4/80+ cells were collected by FACS for RNA-seq. We found that the immune-related pathways, such as myeloid leukocyte migration, positive regulation of cytokine production, leukocyte chemotaxis, regulation of immune effector process, and response to interleukin-1, were downregulated in GFP+F4/80+ cells, as compared with the GFP–F4/80+ cells (Fig. 6A; Supplementary Fig. S6A). GSEA showed that the GFP+F4/80+ cells downregulated the M1 signature (NES = –1.71, P = 0.0085; Fig. 6B), whereas the M2 signature did not change significantly, as compared with GFP–F4/80+ cells (Supplementary Fig. S6B).
Figure 6.
Immunosuppressive activity of double-positive TAMs. A, Pathways that downregulated in GFP+F4/80+ BMDMs, as compared with GFP–F4/80+ BMDMs. B, GSEA plot of GFP+F4/80+ BMDMs compared with GFP–F4/80+ BMDMs. GSEA was performed by the R package clusterProfiler. C, Comparison of IL1α, IL1β, TNFα, and IL10 levels in M1 BMDMs under different coculture conditions. M1, M1 macrophages cultured alone; CD45+GFP+, M1 macrophages with GFP fluorescence; CD45+GFP–, M1 macrophages without GFP fluorescence. Data, mean + SEM. P values were calculated by a two-sided Student t test. D, Comparison of mean fluorescence intensity (MFI) of CD163, CD206, CD276, PD-L1, and PD-L2 in M1 BMDMs under different coculture conditions. Data, mean + SEM. P values are calculated by two-sided Student t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Reg., regulation.
To verify the immunosuppressive activity of the double-positive TAMs in our coculture system, we cocultured M1 BMDMs with GFP+ U251. We stained BMDMs with the CD45 antibody and sorted CD45+GFP+ and CD45+GFP– by FACS. ELISA analysis showed that the levels of IL1α, IL1β, and TNFα were significantly decreased (Student t test, P < 0.01), whereas the level of IL10 was significantly increased (Student t test, P < 0.01) in CD45+GFP+ cells, as compared with CD45+GFP– cells (Fig. 6C). In addition, the mean fluorescence intensity of CD163, CD206, CD276, PD-L1, and PD-L2 were significantly increased (Student t test, P < 0.01) in CD45+GFP+ cells, as compared with CD45+GFP– cells (Fig. 6D). These data suggested that the double-positive TAMs showed the phenotype of M2 macrophage and drove immunosuppression via upregulation of PD-L1, PD-L2, and CD276.
Phagocytic ability of BMDMs is correlated with the infiltration of lymphocytes in GBM patients
To correlate phagocytosis with the infiltration of immune cells in GBM patients, we used the single-cell mass cytometry (CyTOF) data from Friebel and colleagues (30), where the infiltrated immune cells in GBM tissues were profiled by antibody panels. We annotated single cells by cell type–specific protein expression, including CD3+ for T cells, CD64+CD3–CD19– for TAMs or monocytes, CD56+CD3−CD19–CD11b– for NK cells, CD66b+CD11b+ for neutrophils, CD11b+CD1c+ for DCs, CD19+CD3–CD11b– for B cells, and CD19–CD38high for plasma cells (Supplementary Fig. S7A–S7B). TAMs and monocytes were reclustered and further classified into BMDM, microglia, and monocytes by the respective expression of CD49d, CX3CR1, and CCR2 (Supplementary Fig. S7C–S7D).
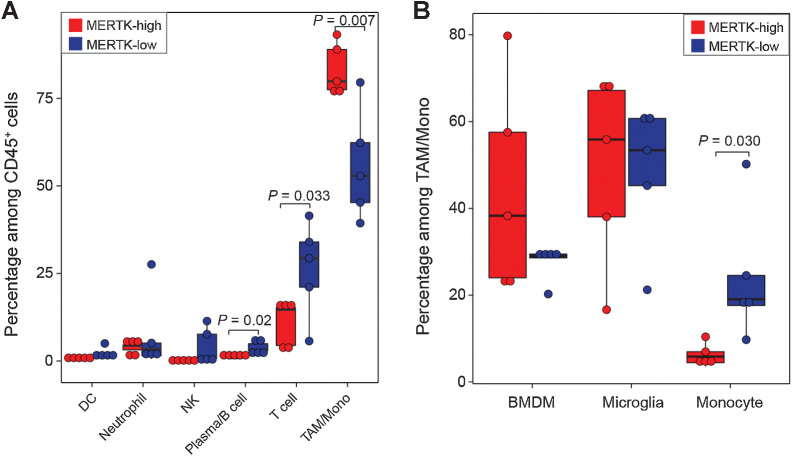
MERTK is a membrane tyrosine kinase and is required for phagocytosis (46, 47). We classified GBM patients into two groups according to the level of MERTK in TAMs. Specifically, ZH802, ZH810, ZH818, ZH791, and ZH784 were classified into the MERTK-high group, whereas ZH794, ZH813, ZH746, ZH761, and ZH816 were classified into the MERTK-low group (Supplementary Table S1). Compared with the MERTK-low group, the MERTK-high group showed increased infiltration of TAMs/monocytes (79.9% vs. 52.9%, P = 0.007) and decreased infiltration of T cells (14.6% vs. 29.3%, P = 0.033) and B/plasma cells (0.87% vs. 3.16%, P = 0.02; Supplementary Fig. S7E; Fig. 7A). We also observed a decreased infiltration of monocytes (5.85% vs. 19%, P = 0.030) and an increased infiltration of BMDMs (38.3% vs. 29%, P = 0.1) in the MERTK-high group (Supplementary Fig. S7F; Fig. 7B).
Figure 7.
BMDM phagocytosis is correlated with the infiltration of lymphocytes. A, Box plots showing the percentage of different immune cells in MERTK-high (n = 5) and MERTK-low (n = 5) GBM patients. P values were calculated by one-sided Student t test. B, Box plots showing the percentage of BMDM, monocyte, and microglia in MERTK-high (n = 5) and MERTK-low (n = 5) GBM patients. P values were calculated by one-sided Student t test.
Discussion
GBM is the most common and lethal malignant brain tumor, which is characterized by systemic immunosuppression and is poorly responsive to current immunotherapies (4, 5). Thus, a thorough understanding of immunosuppression mechanisms is vital to the development of immunotherapy strategies in GBM. In this study, we identified a subset of TAMs that coexpressed macrophage and glioma cell signatures. These double-positive TAMs exhibited enhanced immunosuppressive activity, as compared with normal TAMs. We demonstrated that the double-positive TAMs were formed by phagocytosis of glioma cells and mediated immunosuppression via the transition of M1 macrophages into M2 macrophages, and upregulation of immune-checkpoint proteins CD276, PD-L1, and PD-L2.
Double-positive cells that express macrophage and tumor cell markers have been observed in GBM (12), colon adenocarcinoma (48), melanoma (48), and ovarian cancer (10). These double-positive cells are formed by cell fusion and play roles in tumor invasion, metastasis, and progression. We showed that the double-positive TAMs can also be formed by phagocytosis. Functionally, these double-positive TAMs have enhanced immunosuppressive phenotypes, with decreased T cell–activating signals and increased T-cell inhibiting signals in comparison with normal TAMs. TAMs in the GBM TME include invading BMDMs and brain-resident MG. Interestingly, we observed that double-positive TAMs tended to express BMDM markers in comparison with MG markers (Fig. 2H), indicating that BMDMs may be the main contributors to the phagocytosis of glioma cells. In addition, BMDMs reside in the surrounding region of the vascular and core tumor regions, whereas MG resides in the marginal tumor regions (36). Based on their location, BMDMs have a greater probability of phagocytosing tumor cells. Therefore, we speculate that the tendency of double-positive TAMs to be BMDM-like may be associated with the distribution of BMDMs.
Phagocytosis mediated by macrophages is critical for the activation of both innate and adaptive immune systems (49). However, GBM cells highly express the “don't eat me” signal CD47, which interacts with signal-regulatory protein α (SIRPα) on the macrophages to evade engulfment. Currently, targeting the phagocytosis checkpoint CD47–SIRPα axis is considered an antitumor therapy. However, anti-CD47 alone has limited survival benefits in the murine GBMs (50), of which, the underlying mechanism is still unclear. Our study provided a potential explanation that the phagocytosed tumor cells could endow macrophages with an immunosuppressive phenotype associated with highly expressing immune-checkpoint proteins such as PD-L1, PD-L2, etc., eventually leading to the diminished effect of treatment. Thus, to achieve durable antitumor effects, combining prophagocytosis agents (such as anti-CD47) with immune-checkpoint inhibitors (such as anti–PD-L1) could be a candidate treatment strategy, yet still need more evidence.
While introducing the formation of double-positive macrophages, we recognize that the current study has not fully uncovered the underlying mechanism of how the phagocytosing tumor cells endow macrophages with an immunosuppressive phenotype. A detailed analysis should be further performed in our future study.
In summary, our study reveals the immunosuppressive activity of double-positive TAMs formed by the phagocytosis of glioma cells. These findings provide a deepened understanding of phagocytic macrophage-mediated immunosuppression.
Supplementary Material
Clinical information of GBM patients included in this study
Gene signatures used for ssGSEA
Information of TAMs harboring tumor-derived mutations
Macrophages phagocytose tumor cells.
Supplementary Figures S1-7
Acknowledgments
The authors would like to thank Yangqing Li (School of Basic Medical Sciences, Nanjing Medical University), Danyang Shan (School of Basic Medical Sciences, Nanjing Medical University), and Lang Hu (School of Basic Medical Sciences, Nanjing Medical University) for the culture of GSCs and NSCs, and Yang Chen (Shanghai Institute of Nutrition and Health, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences) for work discussion and image processing. This study was supported by the National Natural Science Foundation of China (Grant Nos. 81572893, 81972358, 91959113, 31771334, and 81970428), the Key Research and Development Program of Jiangsu Province (Grant No. BE2017733), the Basic Research Program of Jiangsu Province (Grant No. BK20180036), Jiangsu Province's Science and Technology Foundation (Grant No. BE2018724), the Major Research Plan of the National Natural Science Foundation of China (Grant No. 91649125), the National Key R&D Program of China (Grant No. 2016YFA0503100), and the Priority Academic Program Development of Jiangsu Higher Education Institutions.
The publication costs of this article were defrayed in part by the payment of publication fees. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Authors' Disclosures
Q. Wang reports nonfinancial support from Oncocare Life Technology (Suzhou) Co., Ltd. outside the submitted work. No disclosures were reported by the other authors.
Authors' Contributions
M. Wu: Conceptualization, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. L. Wu: Investigation, writing–review and editing. W. Wu: Software, investigation. M. Zhu: Investigation. J. Li: Investigation. Z. Wang: Formal analysis. J. Li: Formal analysis. R. Ding: Software. Y. Liang: Software. L. Li: Software. T. Zhang: Formal analysis. B. Huang: Formal analysis. Y. Cai: Formal analysis. K. Li: Formal analysis. L. Li: Software. R. Zhang: Software. B. Hu: Methodology. F. Lin: Methodology. X. Wang: Methodology. S. Zheng: Methodology. J. Chen: Methodology. Y. You: Supervision, funding acquisition. T. Jiang: Supervision, funding acquisition. J. Zhang: Supervision, funding acquisition. H. Chen: Funding acquisition, methodology. Q. Wang: Conceptualization, resources, supervision, funding acquisition, writing–original draft, project administration, writing–review and editing.
References
- 1. Tan AC, Ashley DM, Lopez GY, Malinzak M, Friedman HS, Khasraw M. Management of glioblastoma: state of the art and future directions. CA Cancer J Clin 2020;70:299–312. [DOI] [PubMed] [Google Scholar]
- 2. Luke JJ, Flaherty KT, Ribas A, Long GV. Targeted agents and immunotherapies: optimizing outcomes in melanoma. Nat Rev Clin Oncol 2017;14:463–82. [DOI] [PubMed] [Google Scholar]
- 3. Gettinger SN, Wurtz A, Goldberg SB, Rimm D, Schalper K, Kaech S, et al. Clinical features and management of acquired resistance to PD-1 axis inhibitors in 26 patients with advanced non-small cell lung cancer. J Thorac Oncol 2018;13:831–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jackson CM, Choi J, Lim M. Mechanisms of immunotherapy resistance: lessons from glioblastoma. Nat Immunol 2019;20:1100–9. [DOI] [PubMed] [Google Scholar]
- 5. Lim M, Xia Y, Bettegowda C, Weller M. Current state of immunotherapy for glioblastoma. Nat Rev Clin Oncol 2018;15:422–42. [DOI] [PubMed] [Google Scholar]
- 6. DeNardo DG, Ruffell B. Macrophages as regulators of tumour immunity and immunotherapy. Nat Rev Immunol 2019;19:369–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bowman RL, Klemm F, Akkari L, Pyonteck SM, Sevenich L, Quail DF, et al. Macrophage ontogeny underlies differences in tumor-specific education in brain malignancies. Cell Rep 2016;17:2445–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Muller S, Kohanbash G, Liu SJ, Alvarado B, Carrera D, Bhaduri A, et al. Single-cell profiling of human gliomas reveals macrophage ontogeny as a basis for regional differences in macrophage activation in the tumor microenvironment. Genome Biol 2017;18:234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pinton L, Masetto E, Vettore M, Solito S, Magri S, D'Andolfi M, et al. The immune-suppressive microenvironment of human gliomas depends on the accumulation of bone marrow-derived macrophages in the center of the lesion. J Immunother Cancer 2019;7:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Akhter MZ, Sharawat SK, Kumar V, Kochat V, Equbal Z, Ramakrishnan M, et al. Aggressive serous epithelial ovarian cancer is potentially propagated by EpCAM(+)CD45(+) phenotype. Oncogene 2018;37:2089–103. [DOI] [PubMed] [Google Scholar]
- 11. Gast CE, Silk AD, Zarour L, Riegler L, Burkhart JG, Gustafson KT, et al. Cell fusion potentiates tumor heterogeneity and reveals circulating hybrid cells that correlate with stage and survival. Sci Adv 2018;4:eaat7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cao MF, Chen L, Dang WQ, Zhang XC, Zhang X, Shi Y, et al. Hybrids by tumor-associated macrophages x glioblastoma cells entail nuclear reprogramming and glioblastoma invasion. Cancer Lett 2019;442:445–52. [DOI] [PubMed] [Google Scholar]
- 13. Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for illumina sequence data. Bioinformatics 2014;30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Siren J, Valimaki N, Makinen V. Indexing graphs for path queries with applications in genome research. IEEE/ACM Trans Comput Biol Bioinform 2014;11:375–88. [DOI] [PubMed] [Google Scholar]
- 15. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–30. [DOI] [PubMed] [Google Scholar]
- 16. Qiu X, Hill A, Packer J, Lin D, Ma YA, Trapnell C. Single-cell mRNA quantification and differential analysis with census. Nat Methods 2017;14:309–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics 2013;43:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puram SV, Tirosh I, Parikh AS, Patel AP, Yizhak K, Gillespie S, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell 2017;171:1611–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016;32:2847–9. [DOI] [PubMed] [Google Scholar]
- 21. Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018;36:411–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Welch JD, Kozareva V, Ferreira A, Vanderburg C, Martin C, Macosko EZ. Single-cell multi-omic integration compares and contrasts features of brain cell identity. Cell 2019;177:1873–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H, Durbin R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Talevich E, Shain AH, Botton T, CNVkit BBC.: Genome-Wide copy number detection and visualization from targeted DNA sequencing. PLoS Comput Biol 2016;12:e1004873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nowicka M, Crowell HL, Robinson MD. 2020 cytofWorkflow: CyTOF workflow: differential discovery in high-throughput high-dimensional cytometry datasets. <https://github.com/markrobinsonuzh/cytofWorkflow>. [DOI] [PMC free article] [PubMed]
- 26. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bengtsson H, Ray A, Spellman P, Speed TP. A single-sample method for normalizing and combining full-resolution copy numbers from multiple platforms, labs and analysis methods. Bioinformatics 2009;25:861–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Couturier CP, Ayyadhury S, Le PU, Nadaf J, Monlong J, Riva G, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun 2020;11:3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Friebel E, Kapolou K, Unger S, Nunez NG, Utz S, Rushing EJ, et al. Single-cell mapping of human brain cancer reveals tumor-specific instruction of tissue-invading leukocytes. Cell 2020;181:1626–42. [DOI] [PubMed] [Google Scholar]
- 31. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 2015;1:417–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen WT, Lu A, Craessaerts K, Pavie B, Sala Frigerio C, Corthout N, et al. Spatial transcriptomics and in situ sequencing to study alzheimer's disease. Cell 2020;182:976–91. [DOI] [PubMed] [Google Scholar]
- 33. Moesta AK, Li XY, Smyth MJ. Targeting CD39 in cancer. Nat Rev Immunol 2020;20:739–55. [DOI] [PubMed] [Google Scholar]
- 34. Buchbinder EI, Desai A. CTLA-4 and PD-1 pathways: similarities, differences, and implications of their inhibition. Am J Clin Oncol 2016;39:98–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Andersen BM, Faust Akl C, Wheeler MA, Chiocca EA, Reardon DA, Quintana FJ. Glial and myeloid heterogeneity in the brain tumour microenvironment. Nat Rev Cancer 2021;21:786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Klemm F, Maas RR, Bowman RL, Kornete M, Soukup K, Nassiri S, et al. Interrogation of the microenvironmental landscape in brain tumors reveals disease-specific alterations of immune cells. Cell 2020;181:1643–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sankowski R, Böttcher C, Masuda T, Geirsdottir L, Sagar Sindram E, et al. Mapping microglia states in the human brain through the integration of high-dimensional techniques. Nat Neurosci 2019;22:2098–110. [DOI] [PubMed] [Google Scholar]
- 38. Pombo Antunes AR, Scheyltjens I, Lodi F, Messiaen J, Antoranz A, Duerinck J, et al. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci 2021;24:595–610. [DOI] [PubMed] [Google Scholar]
- 39. Aguilar PS, Baylies MK, Fleissner A, Helming L, Inoue N, Podbilewicz B, et al. Genetic basis of cell-cell fusion mechanisms. Trends Genet 2013;29:427–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Brennan CW, Verhaak RG, McKenna A, Campos B, Noushmehr H, Salama SR, et al. The somatic genomic landscape of glioblastoma. Cell 2013;155:462–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao Z, Zhang KN, Wang Q, Li G, Zeng F, Zhang Y, et al. Chinese glioma genome atlas (CGGA): a comprehensive resource with functional genomic data from Chinese glioma patients. Genomics Proteomics Bioinformatics 2021;19:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and related cytokines in the regulation of inflammation and immunity. Immunity 2019;50:778–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Widodo SS, Dinevska M, Furst LM, Stylli SS, Mantamadiotis T. IL-10 in glioma. Br J Cancer 2021;125:1466–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep 2014;6:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12:252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhou Y, Fei M, Zhang G, Liang WC, Lin W, Wu Y, et al. Blockade of the phagocytic receptor MerTK on tumor-associated macrophages enhances P2×7R-dependent STING activation by tumor-derived cGAMP. Immunity 2020;52:357–73. [DOI] [PubMed] [Google Scholar]
- 47. Arandjelovic S, Ravichandran KS. Phagocytosis of apoptotic cells in homeostasis. Nat Immunol 2015;16:907–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Powell AE, Anderson EC, Davies PS, Silk AD, Pelz C, Impey S, et al. Fusion between intestinal epithelial cells and macrophages in a cancer context results in nuclear reprogramming. Cancer Res 2011;71:1497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Feng M, Jiang W, Kim BYS, Zhang CC, Fu YX, Weissman IL. Phagocytosis checkpoints as new targets for cancer immunotherapy. Nat Rev Cancer 2019;19:568–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. von Roemeling CA, Wang Y, Qie Y, Yuan H, Zhao H, Liu X, et al. Therapeutic modulation of phagocytosis in glioblastoma can activate both innate and adaptive antitumour immunity. Nat Commun 2020;11:1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Clinical information of GBM patients included in this study
Gene signatures used for ssGSEA
Information of TAMs harboring tumor-derived mutations
Macrophages phagocytose tumor cells.
Supplementary Figures S1-7