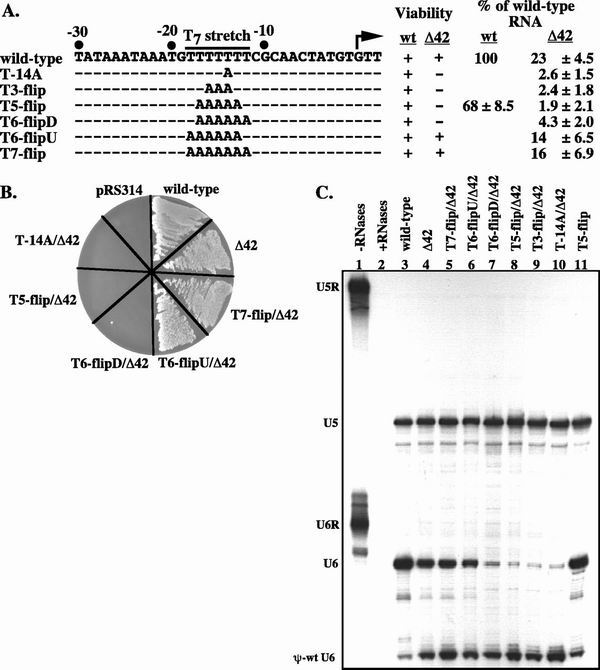
FIG. 4.
Full and partial inversions of the T7 stretch suggest that it acts as a structural element in promoting transcription. (A) Sequence upstream of the transcription start site in wild-type and T7 stretch mutant alleles in the nontemplate strand; the TATA box is located at position −30 to −23. Dashed lines indicate no change from the wild-type sequence. Plus and minus signs indicate viability and inviability, respectively. The amount of U6 RNA present in each strain is designated as the percent of the wild-type (wt) value. Values listed are the averages of at least four experiments and are shown ± standard deviation. (B) Viability test to determine if any of the T7 stretch mutants are synthetically lethal with Δ42. Cells were grown in YEPD followed by plating to 5-FOA, −Trp, to select against the plasmid harboring the pseudo-wild-type U6 allele. The plates were incubated at 30°C for 2 days. pRS314 contains no SNR6 allele and should therefore die on 5-FOA, −Trp. (C) Total cellular RNA from strains containing the indicated SNR6 allele and the pseudo-wild-type U6 allele on separate plasmids was analyzed by RNase protection assay. Samples were run on an 8.3 M urea–6% polyacrylamide gel and exposed to a PhosphorImager screen overnight. U5 RNA is a normalization standard in this experiment. The undigested probes and probes digested in the absence of total cellular RNA are shown in lanes 1 and 2, respectively.