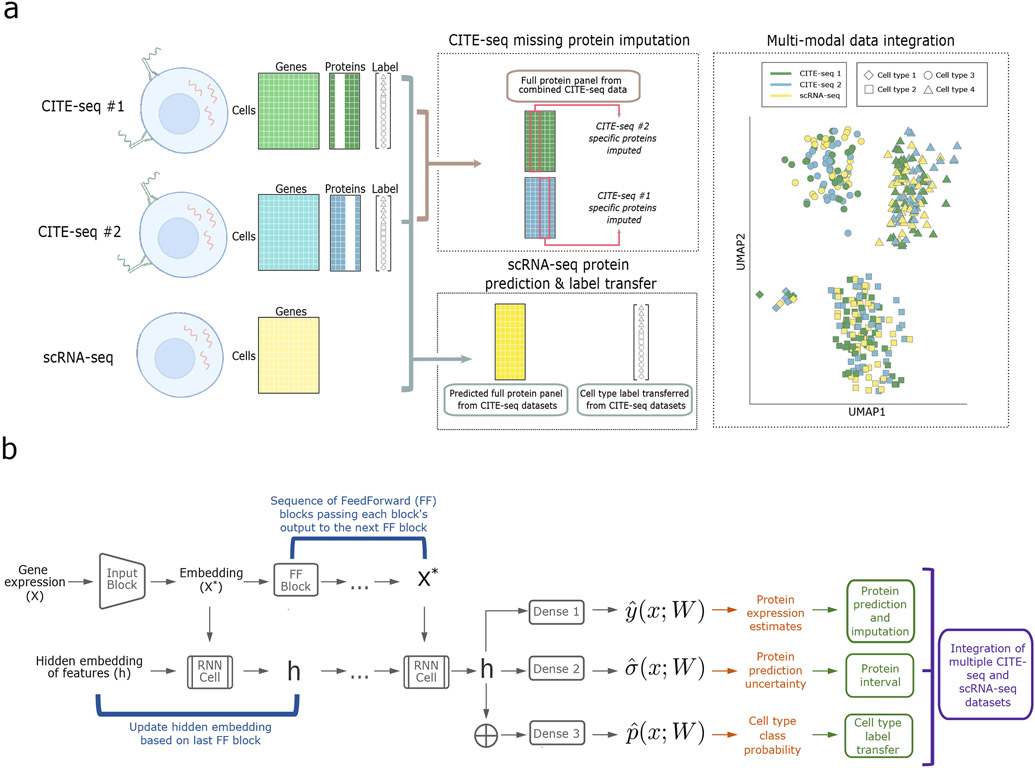
Fig. 1 ∣. Overview of sciPENN.
a, sciPENN is a flexible method which supports completion of multiple CITE-seq references (by imputing missing proteins for each reference) as well as protein expression prediction in an scRNA-seq test set, all in one framework. Simultaneously, sciPENN can transfer cell type labels from a training set to a test set, and can also integrate cells from the multiple datasets into a common latent space. b, sciPENN’s model architecture is comprised by an input block, followed by a sequence of FeedForward blocks interleaved with updates to an internally maintained hidden state updated via an RNN cell. The final hidden state is passed through three dense layers to compute protein predictions, protein prediction bounds, and cell type class probability vectors.