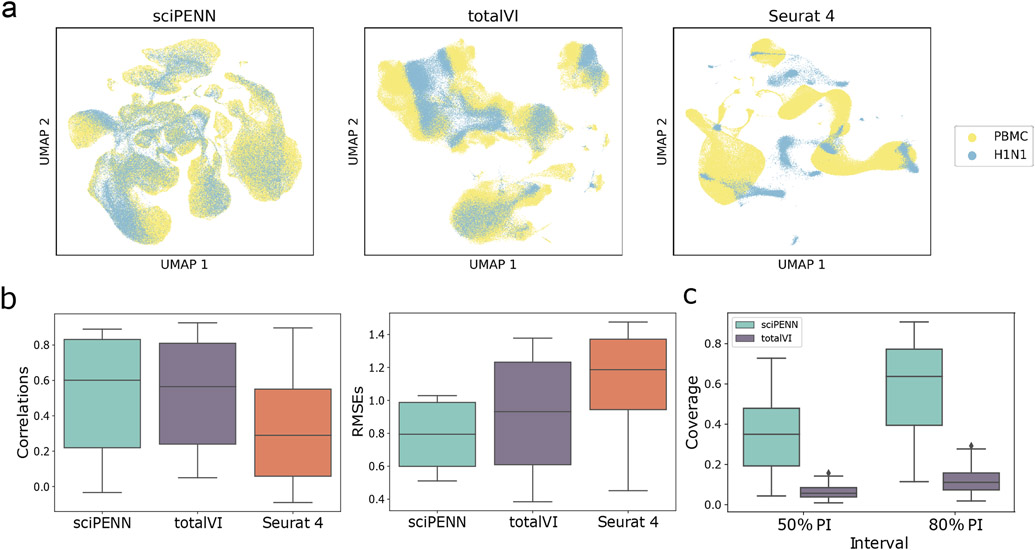
Fig. 5 ∣. Protein expression prediction in the H1N1 dataset using the Seurat 4 PBMC dataset as reference.
a, UMAP Embeddings visualizing the integrated hidden representation of the data, for each method. Each cell is colored according to the dataset from which it was sequenced. b, Box plots that display the correlation (left) and the RMSE (right) between each H1N1 protein’s predicted and true values for each method. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 53,201 cells in the H1N1 dataset and 161,764 cells in the Seurat 4 PBMC dataset. c, Box plots that visualize the empirical test coverage of nominal 50% and 80% PIs per protein computed with sciPENN and totalVI. The lower and upper hinges correspond to the first and third quartiles, and the center refers to the median value. The upper (lower) whiskers extend from the hinge to the largest (smallest) value no further (at most) than 1.5 × interquartile range from the hinge. Results are based on the analysis of 53,201 cells in the H1N1 dataset and 161,764 cells in the Seurat 4 PBMC dataset.