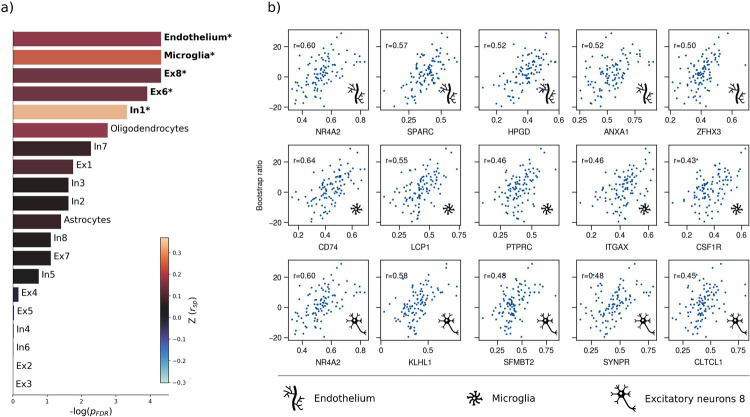
Figure 3. Virtual histology analysis.
The correspondence between MetS effects (bootstrap ratio) and cell type-specific gene expression profiles was examined via an ensemble-based gene category enrichment analysis. a) Barplot displaying spatial correlation results. The bar height displays the significance level. Colors encode the aggregate z-transformed Spearman correlation coefficient relating the Schaefer100-parcellated bootstrap ratio and respective cell population densities. b) Scatter plots illustrating spatial correlations between MetS effects and exemplary cortical gene expression profiles per cell population significantly associated across analyses – i.e., endothelium, microglia and excitatory neurons type 8. Top 5 genes most strongly correlating with the bootstrap ratio map were visualized for each of these cell populations. Icons in the bottom right of each scatter plot indicate the corresponding cell type. A legend explaining the icons is provided at the bottom. First row: endothelium; second row: microglia; third row: excitatory neurons type 8. A corresponding plot illustrating the contextualization of the t-statistic derived from group statistics is shown in supplementary figure S20. Abbreviations: – negative logarithm of the false discovery rate-corrected p-value derived from spatial lag models [38,40]; – Spearman correlation coeffient. – aggregate z-transformed Spearman correlation coefficient.