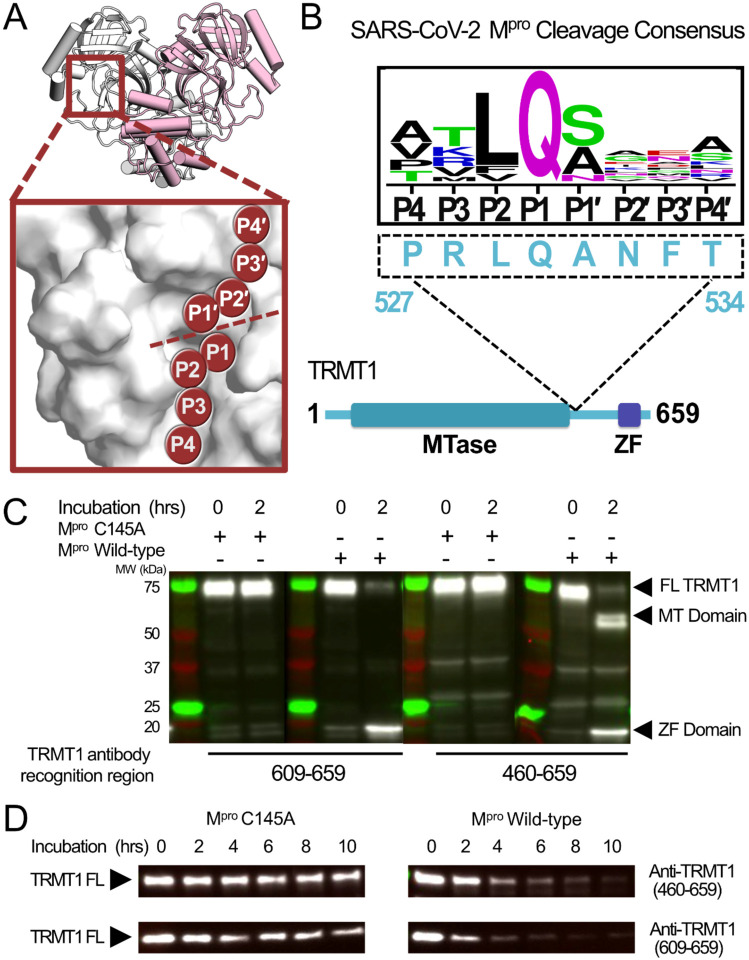
Figure 1.
SARS-CoV-2 Mpro cleaves full-length human TRMT1. A) Overview of the structure of the SARS-CoV-2 Mpro homodimer (PDB 7BB2) with substrate peptide residues (P4-P3-P2-P1-P1′-P2′P3′-P4′) illustrated in the Mpro active site (inset); proteolytic cleavage takes place between substrate residues P1 and P1′ (dotted line). B) The TRMT1(527–534) sequence found in a linker region between the TRMT1 SAM methyltransferase (MTase) and Zinc Finger (ZF) domains is consistent with the SARS-CoV-2 Mpro cleavage consensus sequence. C) Western blots of recombinantly expressed full-length TRMT1 incubated with 10 μM catalytically inactive (Cys145Ala) or active (Wild-type) SARS-CoV-2 Mpro at 37°C. Incubation with WT Mpro results in proteolysis of FL TRMT1 and the appearance of cleavage products corresponding the ZF domain (observed with both antiTRMT1(609–659) and anti-TRMT1(460–659) antibodies) and the MTase domain (observed with only anti-TRMT1(460–659) antibody). D) Western blots of endogenous human TRMT1 in HEK293T cell lysate incubated with 10 μM of either catalytically inactive (Cys145Ala) or active (Wild-type) Mpro at 37°C. Endogenous FL TRMT1 is stable in human cell lysate over the course of a 10-hour incubation with C145A Mpro (left) and is rapidly proteolyzed upon incubation with WT Mpro (right).