Figure 5.
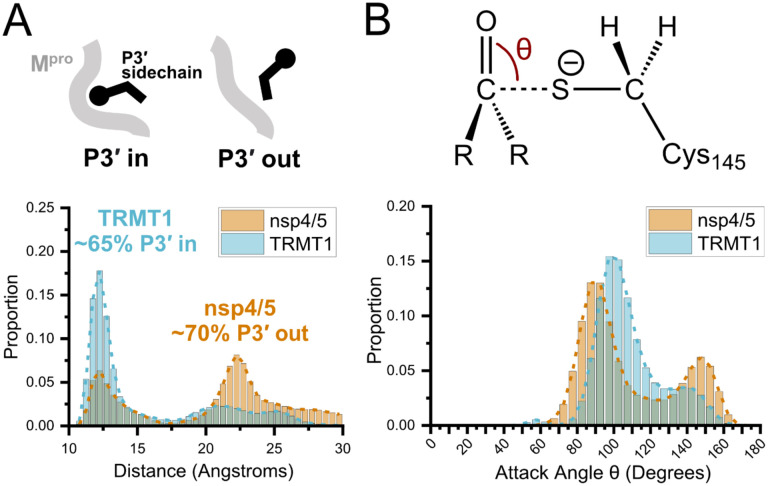
Molecular dynamics (MD) simulations confirm dominant peptide binding conformations and suggest discrimination in cleavage kinetics result catalytic steps that follow initial binding and nucleophilic attack. A) Distribution of the sum of the minimum distance for P3′ Phe residue in nsp4/5 or TRMT1 from three residues (Thr25, Met49, Cys44) which form the S3′ subsite; P3′-in and P3′-out conformations are illustrated above the distribution plot. The much larger proportion of TRMT1 at smaller distances reflects the peptide’s preference for binding in the P3′-in conformation where TRMT1 P3′ Phe occupies the S3′ pocket during the majority of the MD simulation. B) Distribution of the attack angle of the nucleophilic Mpro Cys145 sulfur atom and the substrate carbonyl carbon atom in the to-be-cleaved amide bond (S–C=O angle q, top illustration) during the course of the MD simulation. Although nsp4/5 has a higher proportion of attack angles observed closer to the optimal 90 degrees compared to TRMT1, consistent with faster nsp4/5 cleavage kinetics, this small preference is insufficient to explain the 200-fold faster cleavage kinetics of nsp4/5 observed in experimental proteolysis assays.