Figure 4.
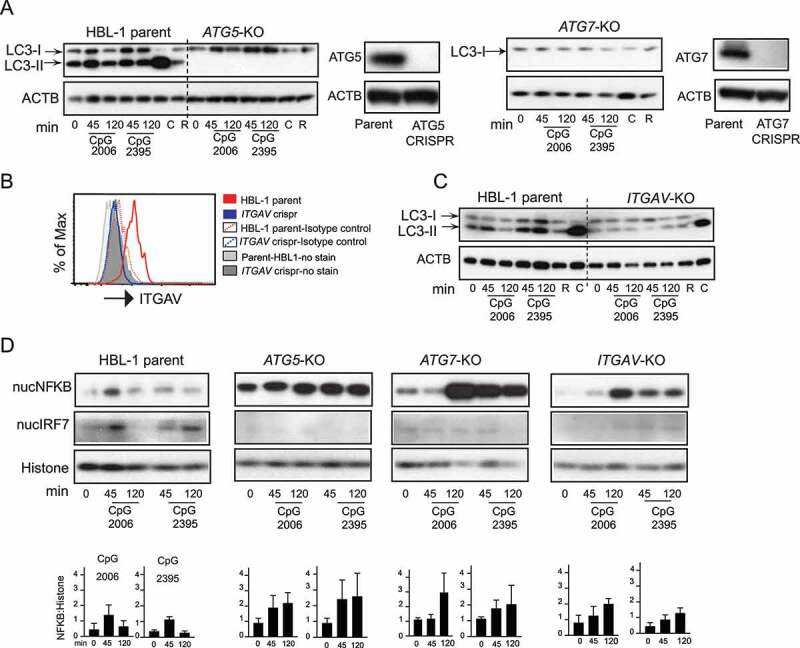
Integrin-dependent autophagy regulates TLR signaling in activated human B cell line. (A) Western blot analysis of LC3 processing or LC3 lipidation in parental HBL-1 cells or cells with CRISPR of autophagy genes (ATG5 or ATG7) labeled as ATG5-KO and ATG7-KO. Parental cells or CRISPR cells were stimulated with either CpG 2006 or CpG 2395 or with autophagy modulators rapamycin (R) or chloroquine (C) for indicated times, and cytoplasmic extracts were used to assess LC3. ACTB is shown as loading control. Western blot analysis of changes in the protein levels of ATG5 or ATG7 in the CRISPR cells compared to parental cells are also shown. (B) Flow cytometry analysis for changes in protein levels of ITGAV integrin after CRISPR of the ITGAV gene. (C) LC3 lipidation in parental HBL-1 cells or HBL-1 cells with CRISPR of ITGAV gene. (D) Nuclear fraction from the stimulated parental cells (HBL-1 parent) or cells with CRISPR of ATG5 gene (ATG5-KO) or ATG7 gene (ATG7-KO) or ITGAV gene (ITGAV-KO) were analyzed for NFKB and IRF7 activation. RELA/p65 for NFKB and IRF7 levels are shown by western blot. Histone is shown as nuclear loading control. Graphs show quantification for NFKB upon either CpG 2006 or CpG 2395 stimulation in relation to loading controls. Quantification is based on densitometry from at least three independent experiments. In all cases, representative blots from at least three independent experiments are shown.
