Figure 2.
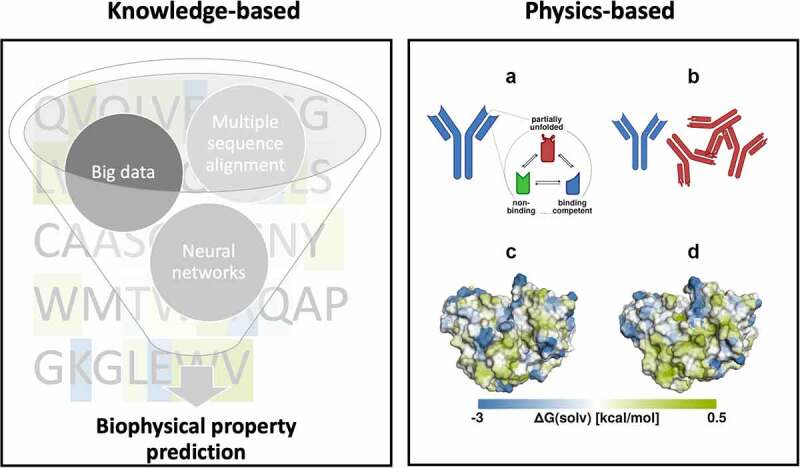
Knowledge and physics-based approaches for characterizing biophysical properties of antibodies. Left panel, knowledge-based: Overview of critical steps for sequence-based in silico prediction of biophysical properties from several thousands of potential hit sequences. Right panel, physics-based: A) The antibody binding interface exists as an ensemble of conformations, which includes binding competent as well as non-binding states. Partially unfolded conformations also exist with a lower probability. B) Different conformations exhibit different properties, where partially unfolded conformations may aggregate which leads to further unfolding. In C) and D), the hydrophobicity profile of two different conformations of the TNF-α binding antibody golimumab is mapped on its molecular surface using localized free energy of hydration. The two conformations show a significantly altered hydrophobicity profile and will therefore most likely interact differently with other hydrophobic molecules.
