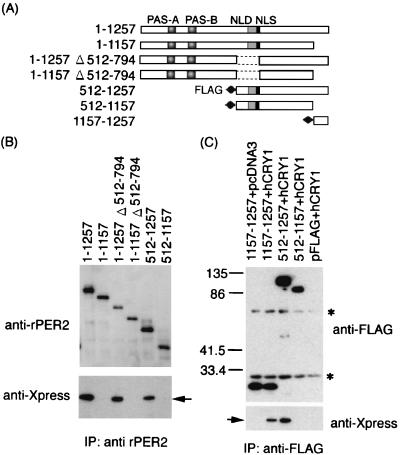
FIG. 5.
Coimmunoprecipitation shows that hCRY1 interacts with rPER2 carboxyl-terminal domain. (A) Schematic representation of rPER2 mutant constructs for coimmunoprecipitation. The positions of rPER2 NLD, PAS-A, and PAS-B are shown by shaded boxes. The rPRE2 NLS is shown as a solid box. The mutants with a deletion at the amino-terminal half (512–1257 and 512–1157) and at the carboxyl-terminal fragment of rPER2 (1157–1257) were tagged with FLAG at their amino-terminal ends. (B) Total lysates from cells coexpressing hCRY1 and truncation mutants of rPER2 were immunoprecipitated (IP) and blotted with anti-rPER2 antibodies (top panel) and were detected using anti-Xpress antibodies (lower panel). Results were similar in replicate experiments. Arrow indicates position of hCRY1. (C) Identification of the CRY1 binding domain of rPER2. The FLAG-tagged truncation mutants of rPER2 proteins (1157–1257, 512–1257, and 512–1157) were expressed with hCRY1. Control was vector pcDNA3 or pFLAG instead of hCRY1 or rPER2 mutants. Lysates from transfected cells were immunoprecipitated (IP) and blotted with anti-FLAG antibodies (top panel) and were visualized using anti-Xpress antibodies (lower panel). The arrow indicates the position of hCRY1. Asterisks indicate nonspecific band corresponding to Ig derived from M2 monoclonal antibodies. Numerals indicate molecular weights in thousands.