Abstract
Background
Sarbecoviruses are a subgenus of Coronaviridae that mostly infect bats with known potential to infect humans (SARS-CoV and SARS-CoV-2). Populations in Southeast Asia, where these viruses are most likely to emerge, have been undersurveyed to date.
Methods
We surveyed communities engaged in extractive industries and bat guano harvesting from rural areas in Myanmar. Participants were screened for exposure to sarbecoviruses, and their interactions with wildlife were evaluated to determine the factors associated with exposure to sarbecoviruses.
Results
Of 693 people screened between July 2017 and February 2020, 12.1% were seropositive for sarbecoviruses. Individuals were significantly more likely to have been exposed to sarbecoviruses if their main livelihood involved working in extractive industries (logging, hunting, or harvesting of forest products; odds ratio [OR] = 2.71, P = 0.019) or had been hunting/slaughtering bats (OR = 6.09, P = 0.020). Exposure to a range of bat and pangolin sarbecoviruses was identified.
Conclusion
Exposure to diverse sarbecoviruses among high-risk human communities provides epidemiologic and immunologic evidence that zoonotic spillover is occurring. These findings inform risk mitigation efforts needed to decrease disease transmission at the bat-human interface, as well as future surveillance efforts warranted to monitor isolated populations for viruses with pandemic potential.
Keywords: Coronavirus, Sarbecovirus, Bat, Zoonotic, Myanmar, SARS-CoV-2
Introduction
The COVID-19 pandemic, which has profoundly altered the health, livelihoods, and economies of the world, highlights the urgent need for a broad surveillance for zoonotic spillover, especially for pathogens with pandemic potential in at-risk communities. The earliest known cases of COVID-19 from December 2019 were geographically centered on the Huanan Seafood Market in Wuhan, China [1], [2], [3]. Live SARS-CoV-2-susceptible mammals were sold at the market in late 2019 and SARS-CoV-2-positive environmental samples were spatially associated with vendors selling live mammals, suggesting zoonotic emergence at the market [3]. SARS-CoV-2, the virus causing COVID-19, is a betacoronavirus in the Sarbecovirus subgenus [4], which is composed of SARS-CoV-1, SARS-CoV-2, numerous bat viruses, and a small number of pangolin viruses [5,6]. The genome of SARS-CoV-2 has a 96.2% similarity to that of a bat SARS-related coronavirus (SARSr-CoV; RaTG13) collected from an insectivorous Chinese horseshoe bat (Rhinolophus affinis) in Yunnan province, more than 1000 miles away from Wuhan, along the border with Myanmar [1]. Rhinolophus spp. bats in Asia, Europe, and Africa are considered the natural reservoirs of sarbecoviruses [7,8]; however, the concentrated numbers of SARSr-CoVs found in South and Southeast Asia make this a region of high significance for surveillance in wildlife. Genetically similar SARSr-CoVs have been found in other species of Rhinolophid bats in South and Southeast Asia, including Rhinolophus pusillus and Rhinolophus malayanus in China [9]; Rhinolophus cornutus in Japan [10]; Rhinolophus shameli in Cambodia [11]; Rhinolophus acuminatus in Thailand [12]; and Rhinolophus malaanus, Rhinolophus pusillus, and Rhinolophus marchalli in Laos [13].
Given the diversity of closely related sarbecoviruses in bats within the Rhinopholus genus in Asia, SARS-CoV-2 could have arisen from a single bat species or resulted from a recombination of SARSr-CoVs among animals in shared habitats. However, the epidemiologic circumstances surrounding the emergence(s) of SARS-CoV-2 and the ecology of potential pre-emergent viruses within the Sarbecovirus genus remain largely unknown. Investigations into sarbecovirus infections in humans with activities that place them in high contact with bats, such as extractive industries, guano harvesting, and hunting, have been insufficient to date [14]. Given the increasing interconnectedness of rural populations, more effort is needed to identify and characterize the currently unrecognized sarbecoviruses that circulate in wildlife and their potential to spill over to people to better understand the origin of SARS-CoV-2 and to evaluate the potential for the next novel CoV to emerge from wildlife.
CoV richness is strongly correlated with bat richness, suggesting that most CoVs will be found in regions where bat diversity is highest. Based on the distribution and diversity of Rhinolophus bats, Southeast Asia, including Myanmar, Cambodia, Laos, and Thailand, along with Southeastern China, are considered hotspots for sarbecoviruses of potential zoonotic concern [15]. This is supported by the number of CoVs, which have been discovered in China [16], Thailand [17], Cambodia, Lao People's Democratic Republic [18], and Myanmar [19]. The risk for zoonotic viral disease presence and emergence in humans also increases in geographic areas with higher mammal diversity, where previously pristine forests have been recently deforested [20]. This makes the areas of Southeast Asia, which support large, intact natural habitats and have ongoing ecosystem fragmentation, at high risk for disease emergence [21].
Myanmar has taken a proactive approach to diseases emerging from wildlife, partnering with the international community to conduct “upstream” surveillance of viral pathogens in both wildlife and high-risk communities for wildlife contact. In response to the COVID-19 pandemic, several ongoing human surveillance programs were used to evaluate the exposure to sarbecoviruses before the first recognized cases of SARS-CoV-2 in the country and to better understand the types of human behaviors, geographic regions, and potential species implicated in the spillover of sarbecoviruses from wildlife to humans in this important ecological region for CoV emergence.
Methods
Study population and procedures
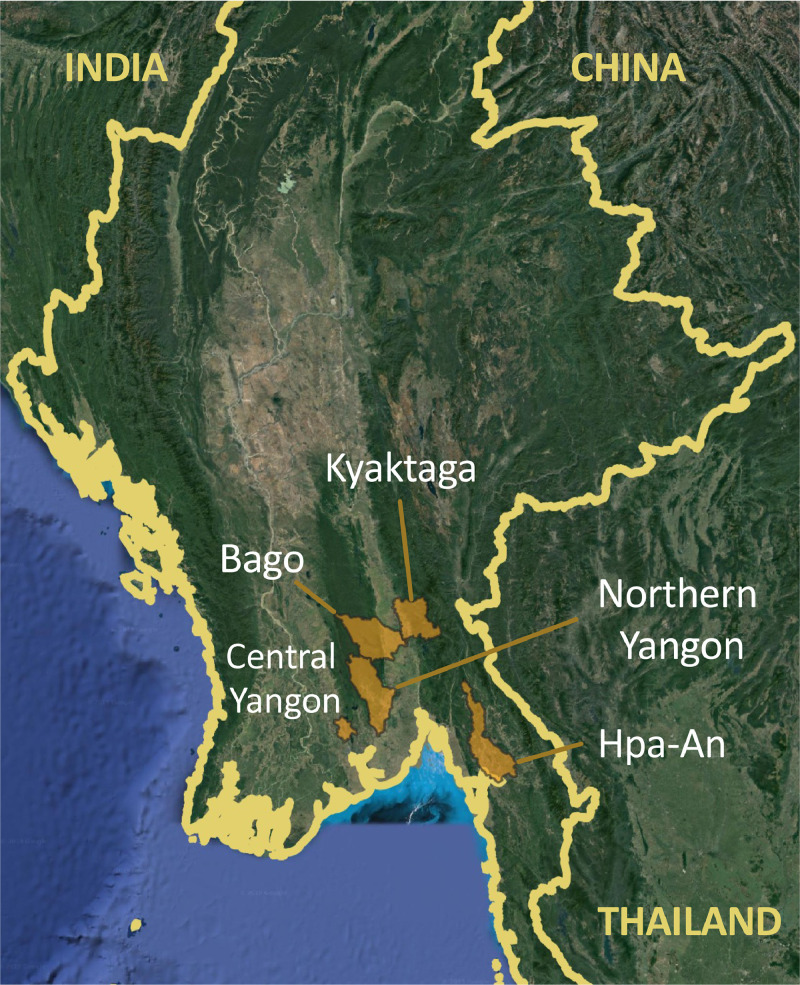
Human subjects were enrolled in this research through three surveillance studies between July 2017 and February 2020 (before the first reported case of SARS-CoV-2 in Myanmar) (Figure 1 ). Written informed consent was obtained from all participants in accordance with the University of California, Davis and Department of Medical Research, Myanmar approved protocols. As part of a United States-Myanmar partnership funded by the National Institutes of Health Fogarty International Center (NIH) to investigate the epidemiology of zoonotic viruses in key biodiversity areas of Myanmar, healthy people (N = 86) were enrolled from five elephant logging camps in the Yenwe Forest Reserve. Myanmar still uses the traditional method of elephant logging for timber harvest and as a result, has a large network of communities where loggers live together in temporary villages on the forest edges with their families and, occasionally, migrant laborers. Cases of acute febrile illness (N = 144) were enrolled from the closest local hospitals to the elephant camps. As part of the United States Agency for International Development Emerging Pandemic Threats PREDICT project, healthy people were enrolled in the study from two regions believed to have high levels of contact with bats, villages from the Hpa-an region (N = 169) and rural Northern Yangon villages (N = 198). As part of the dengue surveillance program conducted by the Department of Medical Research, cases of dengue hemorrhagic fever (N = 98) enrolled at Yangon Children's Hospital were included in the study as an urban non-bat-exposed control group. For the confirmation of assay performance as a validation step, samples from reverse transcription-polymerase chain reaction (RT-PCR)-confirmed cases of COVID-19 (n = 28) during the first COVID-19 wave in Yangon, Myanmar were collected serially at 7-day intervals for 4 weeks after the confirmation of infection status were used. For the NIH and PREDICT studies, a serum sample was collected from all study participants, and a behavioral questionnaire was administered to assess contact with wild animals. Efforts were taken to minimize recall bias by framing all questions in two separate time frames, both “in their lifetime” and “within the past year”. The questions were also repeated in different formats throughout the survey asking about wildlife exposure, both within the context of hunting (which could have legal consequences) and outside the context of hunting. Children enrolled from the city of Yangon were not administered a questionnaire because they were enrolled as part of an acute dengue surveillance program from an urban area of the study. Given the urban nature of this community in Central Yangon, they were considered to not be engaged in high-risk activities involving bats or extractive industries. Bats are not known to be widely distributed within the central metropolitan area of Yangon, and extractive industries are also not located there.
Figure 1.
Locations of human communities within Myanmar.
Diagnostic assays
SARS-CoV-2 surrogate virus neutralization assays
All specimens were tested using a commercially available SARS-CoV-2 surrogate virus neutralization (sVNT) assay that detects the total immunodominant neutralizing antibodies (NAbs) targeting the viral spike protein receptor-binding domain (RBD) in an isotype- and species-independent manner (cPass SARS-CoV-2 Neutralization Antibody Detection Kit, Genscript, Inc). The assay is based on antibody-mediated blockage of the interaction between the human angiotensin-converting enzyme 2 (ACE2) receptor protein and the receptor-binding domain. This assay was validated with two cohorts of patients with COVID-19 in two different countries, achieved 99.9% specificity and 95-100% sensitivity [22] for SARS-CoV-2, and has been shown to not cross-react to other circulating human CoVs, including SARS-CoV. This assay has not been validated for specificity against phylogenetically related sarbecoviruses not yet known to infect humans; therefore, positive cPass results were interpreted as a sample having seropositivity to a virus strain within the Sarbecovirus subgenus. The samples were tested in duplicate according to the manufacturer's instructions at the Department of Medical Research in Myanmar. The cut-off value for the cPass SARS-CoV-2 Neutralizing Antibody Detection Kit is 30% signal inhibition. The percent signal inhibition for the detection of NAbs was calculated as:
Multiplex sVNT assay
A subset of sVNT cPass positive specimens (all positive specimens collected through the NIH surveillance project) was tested for multiple sarbecoviruses using a bead-based multiplex system. The assay measures RBD, targeting NAbs. The assay determines the extent to which NAbs block the interaction between AviTag-biotinylated RBD proteins coated on Luminex microspheres and human ACE2 (calculated as % inhibition). The RBDs included in this study were from (A) clade 2 sarbecoviruses: SARS-CoV-2 Ancestral (NC_045512), SARS-CoV-2 variants of concern (VOCs) (VOCs Alpha (EPI_ISL_2245907), Beta (EPI_ISL_2372356), Lambda (EPI_ISL_3320902), Gamma (EPI_ISL_8939421), Delta (EPI_ISL_5020183), Delta Plus (EPI_ISL_12386865), Mu (EPI_ISL_2897550), Omicron BA.1 (EPI_ISL_7456451), Omicron BA.2 (EPI_ISL_13019463), bat CoV BANAL-52 (MZ937000), Bat CoV BANAL-236 (MZ937003), pangolin CoV GD-1 (EPI_ISL_410721), bat CoV RaTG13 (MN996532), and pangolin CoV GX-P5L (EPI_ISL_410540) and (B) Clade-1 sarbecoviruses: SARS-CoV-1 (AY278488) and bat CoV WIV-1 (KF367457), bat CoV Rs2018B (MK211376), bat CoV LYRa11 (KF569996), and bat CoV RsSHC014 (KC881005). Biotinylated RBD proteins were produced as described previously [23]. Multiplex sVNTs were established as previously described [24]. Briefly, AviTag-biotinylated RBD proteins were coated on MagPlex-Avidin microspheres (Luminex) at 5 μg per 1 million beads. RBD-coated beads (600 per antigen) were preincubated with a testing serum final dilution of 1:20, 1:80, 1:320, 1:1280) for 15 minutes at 37°C with agitation, followed by an addition of 50 μl of phycoerythrin-conjugated human ACE2 (2 mg/ml; Genscript) and incubated for an additional 15 minutes at 37°C, with agitation. After two washes with 1% bovine serum albumin in phosphate buffered saline, the final readings were acquired using the MAGPIX system (Luminex), following the manufacturer's instruction. The cut-off values were set at 30% inhibitation, as described previously for cPass sVNTs.
SARS-CoV-2 plaque reduction neutralization assays
Human sera were thawed at 37°C and 30 µl was heated in a water bath for 30 minutes at 56°C to inactivate the complement proteins. The serum was diluted four-fold with virus diluent consisting of phosphate buffered saline and 1% fetal bovine serum. Then, samples were serially two-fold diluted 11 times for a dynamic range of 1:4 to 1:4096. An equal volume of virus diluent containing 80 plaque-forming units of SARS-CoV-2 was added to each antibody dilution and a no-antibody control consisting of virus diluent only, resulting in a final dynamic range of 1:8-1:8192 with one no-antibody control. The virus strain used was SARS-CoV-2/human/USA/CA-CZB-59 × 002/2020 (GenBank #MT394528), which was isolated from a patient in 2020 in Northern California and passaged once in Vero E6 cells (provided by Dr. Christopher Miller, University of California, Davis). To generate stocks for the plaque reduction neutralization assay (PRNT), SARS-CoV-2 was passaged one additional time in Vero E6 cells to achieve a titer of 2.2 × 107 plaque-forming units/ml. Single-use virus aliquots were stored at -80°C. The antibody-virus dilution series were incubated for 1 hour at 37°C, after which they were applied to confluent Vero CCL-81 cells in single replicate and incubated for 1 hour at 5% CO2 and 37°C in a humidified incubator. The cell monolayers were overlaid with 0.5% agarose dissolved in Dulbecco minimal essential medium with 5% fetal bovine serum and 1 × antibiotic-antimycotic (Fisher Scientific, Waltham, MA, USA) and incubated for 3 days at 5% CO2 and 37°C in a humidified incubator. Cells were fixed for >30 minutes with 4% formaldehyde, then agarose plugs were removed. Cells were stained with 0.05% crystal violet in 20% ethanol for 10 minutes, then rinsed three times with water. The plates were inverted to dry completely, and the number of plaques in each well was counted. The neutralization titer is defined as the reciprocal of the dilution for which fewer than 20% of plaques were detected versus the no-antibody control (>80% neutralization).
Conventional RT-PCR assays
All specimens collected as part of the NIH and PREDICT projects were tested for CoVs using two separate conventional RT-PCR assays. RNA was extracted using Direct-zol RNA kits (Zymo Research, Inc.), according to the manufacturer's instructions. RNA was reverse transcribed into cDNA using SuperScript III (Invitrogen), according to the manufacturer's instructions. Two broadly reactive consensus polymerase chain reaction assays targeting partial and nonoverlapping regions of the CoV ORF1b (containing the RNA-dependent RNA polymerase) were used [25,26]. Bands of the expected size were excised from 1% agarose, cloned into a StrataClone polymerase chain reaction cloning vector, sequenced using Sanger sequencing and compared with available nucleotide sequences in GenBank to confirm the identity.
Statistical analyses
To evaluate associations between human demographic and animal contact risk factors, all covariates were first evaluated for correlation to assess potential confounding. Pearson's chi-square tests were used to determine associations between seropositivity for sarbecoviruses and high-risk human-animal contact behaviors, such as hunting and slaughtering animals, bat guano harvesting, and other contact with wildlife or livelihoods. Statistical tests were considered significant at the level of P-value <0.05. Mixed-effects multivariable logistic regression models were used to assess the association between high-risk wild animal contact behaviors and occupational risk factors that were significant in the bivariate analysis. Township was evaluated as a random variable to account for potential unmeasured risk factors varying by geographic location. Variables were included if they significantly improved the model fit based on the likelihood ratio test (P < 0.1) compared with a model without that variable. Variables were retained in the model if they improved the fit while minimizing the Akaike information criterion. All statistical analyses were performed using STATA/MP 16.1 (StataCorp, College Station, TX, USA).
Results
We identified previous exposure to sarbecoviruses among study participants but did not detect any active infections wherein all participants tested negative for CoVs by consensus RT-PCR or were a confirmed active dengue virus case within the city before the documented emergence of SARS-CoV-2. Among study participants, 12.1% (84/693) were seropositive for sarbecoviruses using the sVNT cPass assay (Table 1 ). Seroexposure was detected among all rural communities and there were no individuals from the urban Central Yangon communities that were seropositive for sarbecoviruses (Figure 1). PRNTs for SARS-CoV-2 performed on seropositive individuals were all negative. A small fraction of study participants was collected during early 2020 before the documented first cases of SARS-CoV-2 in the country. Persons aged older than 20 years were significantly more likely to be seropositive for sarbecoviruses by sVNT (odds ratio [OR] = 2.9, P = 0.009) than those aged younger than 20 years, indicating a higher potential for exposure over time.
Table 1.
Exposure to sarbecoviruses by demographic characteristics among Myanmar communities observed from 2017-2020.⁎
| Characteristic | No. positive | No. negative | Period prevalence (95% confidence interval) | P-value |
|---|---|---|---|---|
| Age | ||||
| <20 years | 7 | 139 | 4.79 (1.95-9.62) | P = 0.002 |
| >20 years | 67 | 457 | 12.79 (10.04-15.95) | * |
| Sex | ||||
| Male | 37 | 317 | 10.45 (7.47-14.11) | P <0.000 |
| Female | 37 | 279 | 11.7 (8.37-15.78) | * |
| Unknowna | 10 | 13 | 43.48 (23.19-65.50) | * |
| Township | ||||
| Bago | 25 | 96 | 20.66 (13.84-28.97) | P <0.000 |
| Hpa-An | 10 | 159 | 5.92 (2.87-10.61) | * |
| Kyauktaga | 14 | 72 | 16.27 (9.20-25.80) | * |
| Central Yangon | 0 | 96 | 0.00 (0.00-3.77) | * |
| North Yangon | 26 | 173 | 12.6 (8.34-18.07) | * |
| Overall | 84 | 609 | 12.12 (9.78-14.78) | * |
Human subjects were enrolled before the first reported case of SARS-CoV-2 in Myanmar.
In the multivariable analysis, individuals who reported that their main livelihood involved extractive industries, including logging, hunting, or harvesting of forest products, were significantly more likely to have been exposed to sarbecoviruses as indicated by seropositivity on the sVNT cPass assay (OR = 2.71, P = 0.019; Table 2 ) compared with those involved in other occupations. When evaluating the specific animal taxa exposure and associated human behaviors, only activities involving bats remained significant in the mixed-effects multivariable logistic regression. Individuals who reported hunting or slaughtering bats in their lifetime were significantly more likely to have been exposed to sarbecoviruses as indicated by seropositivity on the sVNT cPass assay (OR = 6.09, P = 0.020; Table 2) than individuals not reporting these activities. The final mixed-effects multivariable model included hunting/slaughtering bats, extractive industry occupation, and age as fixed effects and township as a random effect (Hosmer-Lemeshow test for logit model without a random effect X 2 = 0.4; P = 0.530). Including township as a random effect significantly improved the model fit (likelihood ratio test vs logistic model: X2 = 13.72; P <0.001).
Table 2.
Distribution of seropositivity to sarbecoviruses among persons exposed and unexposed to wild animals through livelihood and occupational activities in Myanmar.
| Risk factor | Exposed no. persons seropositive/no. tested (%) | Unexposed no. persons seropositive/no. tested (%) | Bivariate |
Multivariable adjusteda |
||
|---|---|---|---|---|---|---|
| ORb | P-value | OR | P-value | |||
| Wildlife hunted or slaughtered | ||||||
| Bat | 3/8 (37.5)c | 71/619 (11.5) | 4.63 | 0.039 | 6.09 | 0.020 |
| Rodent | 6/27 (22.2) | 68/504 (13.5) | 1.83 | 0.208 | NC | NC |
| Primate | 3/20 (15.0) | 71/607 (11.7) | 1.33 | 0.653 | NC | NC |
| Carnivore | 1/16 (6.3) | 73/611 (11.9) | 0.49 | 0.495 | NC | NC |
| Pangolin | 1/14 (7.1) | 73/613 (14.2) | 0.57 | 0.490 | NC | NC |
| Ungulate | 10/38 (26.3) | 64/589 (10.9) | 2.93 | 0.006 | NS | NS |
| All wildlife | 16/69 (23.2) | 58/558 (10.4) | 2.60 | 0.003 | NS | NS |
| Wildlife or excrement from wildlife in house | ||||||
| Bat | 0/1 (0.0) | 1/553 (0.18) | 1.0 | NA | NC | NC |
| Rodent | 25/178 (14.0) | 49/353 (13.9) | 1.01 | 0.959 | NC | NC |
| Primate | 0/15 (0.0) | 74/516 (14.3) | 1.0 | N/A | NC | NC |
| Carnivore | 0/0 (0.0) | 74/531 (13.9) | 1.0 | N/A | NC | NC |
| Pangolin | 0/1 (0.0) | 74/626 (11.8) | 1.0 | N/A | NC | NC |
| Ungulate | 1/17 (5.9) | 73/610 (12.0) | 0.46 | 0.454 | NC | NC |
| All wildlife | 26/199 (13.1) | 48/428 (11.2) | 1.19 | 0.504 | NC | NC |
| Occupation | ||||||
| Extractive industry | 15/45 (33.3) | 69/648 (10.6) | 4.12 | 0.000 | 2.71 | 0.019 |
| Crop production | 20/135 (14.8) | 54/535 (10.1) | 1.59 | 0.120 | NC | NC |
| Animal production | 17/160 (10.6) | 57/494 (11.5) | 0.82 | 0.495 | NC | NC |
| Zoo/animal healthcare | 6/38 (15.8) | 68/632 (10.8) | 1.56 | 0.340 | NC | NC |
| Wild animal trade/market | 0/8 (0.0) | 74/662 (11.2) | 1.0 | N/A | NC | NC |
| Dependent | 8/134 (6.0) | 66/536 (12.3) | 0.45 | 0.041 | NS | NS |
| Migrant laborer | 4/32 (12.5) | 70/638 (11.0) | 1.16 | 0.788 | NC | NC |
Final mixed-effects multivariable model included “Bats hunted or slaughtered”, “Extractive Industry Occupation”, and “Age” as fixed effects and “Township” as a random effect.
OR, odds ratio; NC, not calculated; NS, not significant on multivariable analyses.
Differences in n values were a result of questions being voluntary and people were allowed to skip a question if they felt uncomfortable answering it.
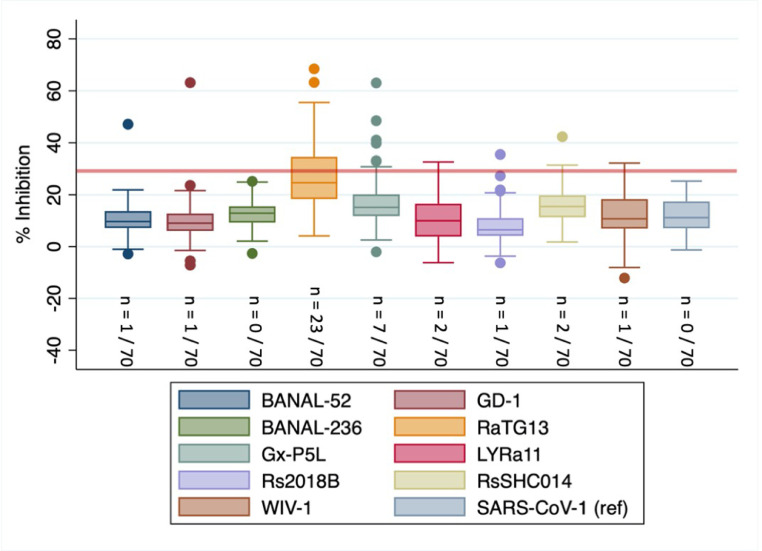
People who were enrolled in the study through the NIH surveillance project in the elephant logging camps and associated clinic catchment areas were also significantly more likely to be exposed to sarbecoviruses (OR = 3.31, P <0.001) than those enrolled through other surveillance programs in different regions of the country. A subset of specimens (n = 70) from this study population was tested using a multiplex sVNT system to further investigate the diversity of sarbecovirus exposures occurring in this population. Preliminary evidence of exposure to a range of sarbecoviruses not yet known to infect humans were detected (Figure 2 ), including 32.8% (23/70) to RaTG13, 1.4% (1/70) to BANAL52, 2.9% (2/70) to LYRa11, 1.4% (1/70) to Rs2018B, 2.9% (2/70) to RsSHC014, and 1.4% (1/70) to WIV bat sarbecoviruses, 10% (7/70) to GxP5L, and 1.4% (1/70) to GD1 pangolin viruses. Low levels of exposure to SARS-CoV-2 VOCs were also detected, including 1.4% (1/70) to Delta, 4.3% (3/70) to Lambda, 2.9% (2/70) to Gamma, 2.9% (2/70) to Omicron BA.1, and 1.4% (1/70) to Mmicron BA.2 at the 30% inhibition cut-off. When evaluating the distributions of percent inhibition values by virus across all individuals tested, only RaTG13 had an interquartile range extending above the designated cut-off value of 30%. All individuals were significantly more likely to have antibodies against sarbecoviruses not yet known to infect humans than sarbecoviruses now known to circulate in humans (i.e., SARS-CoV and SARS-CoV-2 VOCs) (P <0.000) and the detection of pre-emergent sarbecoviruses was not significantly correlated with the detection of sarbecoviruses already documented to have emerged (R2 = 0.099, P = 0.420). People living or working in extractive industry logging camps were significantly more likely to have antibodies against bat CoV RaTG13 (OR = 2.76, P = 0.050) than those reporting other occupations. One individual within the subset of specimens who was evaluated using the multiplex sVNT system self-reported as being a pangolin hunter. This individual was seropositive for the pangolin sarbecovirus GxP5L.
Figure 2.
Exposure to human angiotensin-converting enzyme 2-binding sarbecoviruses not yet known to infect humans among Myanmar extractive industry workers.
*Greater than 30% inhibition of receptor-binding domain binding to human angiotensin-converting enzyme 2 is considered positive.
Discussion
Our findings demonstrate exposure to diverse sarbecoviruses not yet known to infect humans in people exposed to wildlife in Myanmar. The exposure patterns that positively correlated with seropositivity included extractive industries and bat contact, indicating a likely zoonotic transmission through animal contact as opposed to human-to-human transmission. The remote nature of these zoonotic exposure events or the absence of human-to-human transmission could explain why many of these viruses (i.e., Gx-P5L, RsSHCo14, BANAL-52, B21065, LYRa11, WIV-1, GD-1, RaTG13, Rs2018B) are not yet recognized by the global community as having naturally infected humans. Our findings underpin the critical importance of continued surveillance at the rural wildlife-human interface in Southeast Asia, where some of the highest levels of known mammalian diversity exist and where future emergence of zoonotic diseases is likely.
The high relative sarbecovirus seropositivity rate in people engaged in extractive industries highlights the importance of vigilance and access to infectious disease diagnostics in these communities. Our findings are consistent with other studies, which have noted a high risk for infection with zoonotic diseases among people engaged in extractive industries worldwide [27], including Myanmar [28]. The extractive forest industries bring people into contact with wildlife through bushmeat hunting and proximity to the forest, bringing new opportunities for exposure to zoonotic viruses [29]. Given their remote nature, people in these industries often live and work in underserved regions with weak public health and disease prevention infrastructure. Extractive industries, particularly logging, can further contribute to zoonotic disease emergence by altering wildlife habitats and forcing movement of animal populations to more human-dominated areas. Consequently, extractive industry workers and their surrounding communities are especially at risk for emerging infectious diseases.
Direct contact with bats through hunting or slaughtering was a significant risk factor for exposure to sarbecoviruses in our study populations. These findings are consistent with known sarbecoviruses being largely of bat origin. Similar studies conducted in neighboring China have found serological evidence of likely human infection by bat SARSr-CoVs or potentially related viruses in humans living near caves with a known high diversity of bat SARSr-CoVs [14]. Modeling efforts have also predicted a high richness of SARSr-CoV bat host species, as well as bat-human overlap in eastern Myanmar, increasing spillover risk [30]. Mammalian species in decline due to the loss of habitat quality or exploitation have shared more zoonotic viruses with people [31]. Thus, understanding the ecosystem changes that drive bat redistribution after habitat loss and the occupations that bring people and bats into close contact are important for informing public health mitigation measures.
We detected an sVNT antibody reactive to SARS-CoV-2 that did not neutralize SARS-CoV-2 in PRNT assays, indicating that the study participants were likely exposed to other virus strains within the Sarbecovirus genus and not SARS-CoV-2. The most likely explanation is that our findings represent cross-reactivity with closely related known or yet to be discovered sarbecoviruses, further highlighting the need for human and wildlife surveillance in this region. We note that the individuals in our study communities were more likely to have antibodies against sarbecoviruses not yet reported in humans than to sarbecoviruses now known to circulate in humans (i.e., SARS-CoV-2 and SARS-CoV). We know very little about the clinical course of SARS-CoV-2 among isolated human communities with high levels of previous sarbecovirus exposure. An alternate hypothesis is that prior to a zoonotic transmission event from a market in the urban population of Wuhan, SARS-CoV-2 emerged in rural populations with pre-existing pan-sarbecovirus immunity; therefore, the cases were mild or asymptomatic, and stuttering chains of transmission were not detected through existing public health surveillance. This is, however, less likely given the number of documented cases of SARS-CoV-2 in these regions during subsequent stages of the pandemic during 2021-2022.
Antibodies against RaTG13 were the most identified antibodies among all sarbecovirus exposures identified in this study. RaTG13 is the closest known relative to SARS-CoV-2, previously identified in bats. Structural and molecular analysis of the RBD of RaTG13 shows binding affinity to the human receptor ACE2, as well as ACE2 orthologs in 24 other species, providing proof of concept that RaTG13 has the potential to infect humans [32]. Our data suggest that RaTG13 is the sarbecovirus that is most frequently spilling over to people in our study regions. Because RaTG13 was first discovered in a cave in Yunnan, China, along the border with Myanmar, this is not unexpected.
Exposure to sarbecoviruses from a diverse animal origin among biologically plausible high-risk communities, such as forest extractive industry workers and people in contact with bats, provides both epidemiologic and immunologic evidence that zoonotic spillover is occurring in these communities, providing further evidence for the natural origins of SARS-CoV-2 and potential emergence pathways for SARSr-CoVs. Given the worldwide impact of the SARS-CoV-2 pandemic, a better understanding of the transmission mechanisms and specific high-risk behaviors, as well as the impact that pre-existing immunity to sarbecoviruses may have on future SARS-CoV-2 variant exposure in these isolated populations is urgently needed.
Declaration of competing interest
The authors have no competing interests to declare.
Acknowledgments
Funding
The research reported in this publication was supported by the Fogarty International Center of the National Institutes of Health under grant no. K01TW010279, the National Institute of Allergy and Infectious Diseases under grant no. 1U01AI151814-0, the United States Agency for International Development Emerging Pandemic Threats PREDICT project (cooperative agreement no. GHN-A-OO-09-00010-00), the National Science Foundation under grant no. 2109860 and the National Medical Research Council Singapore under grant no. (MOH-000535/MOH-OFYIRG19nov-0002 and OFLCG19May-0034).
Ethical approval
This study was reviewed and approved by the University of California Davis Institutional Review Board (#889159-9 and 804522) and the Department of Medical Research Myanmar's ethics review committee (#012816, #002617, and #2020-119).
Acknowledgments
The authors thank the Department of Medical Research, the Livestock Breeding and Veterinary Department, the Myanmar Timber Enterprise and the Forest Department of the Republic of the Union of Myanmar for their support and facilitation of this research.
Author contributions
Tierra Smiley Evans: conceptualization, methodology, formal analysis, resources, data curation, writing – original draft, visualization, funding acquisition; Chee Wah Tan: methodology, investigation; Ohnmar Aung: methodology, investigation; Sabai Phyu: investigation; Htin Lin: investigation; Lark Coffey: methodology, investigation, writing-review & editing; Aung Than Toe: project administration, investigation; Pyaephyo Aung: project administration, investigation; Tin Htun Aung: investigation; Nyein Thu Aung: investigation; Christopher Weiss: investigation, writing – review & editing; Kyaw Zin Thant: Supervision, writing – review & editing; Zaw Than Htun: supervision; Suzan Murray: Supervision, project administration, funding acquisition; Linfa Wang: supervision, resources, funding acquisition; Christine Johnson: writing – review & editing, supervision, funding acquisition; Hlaing Myat Thu: supervision, project administration, writing – review & editing.
References
- 1.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Worobey M, Levy JI, Malpica Serrano L, Crits-Christoph A, Pekar JE, Goldstein SA, Rasmussen AL, Kraemer MUG, Newman C, Koopmans MPG, Suchard MA, Wertheim JO, Lemey P, Robertson DL, Garry RF, Holmes EC, Rambaut A, Andersen KG. The Huanan Seafood Wholesale Market in Wuhan was the early epicenter of the COVID-19 pandemic. Science. 2022;377:951–959. doi: 10.1126/science.abp8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coronaviridae Study Group of the International Committee on Taxonomy of Viruses The species severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol. 2020;5:536–544. doi: 10.1038/s41564-020-0695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Shi Z, Yu M, Ren W, Smith C, Epstein JH, Wang H, Crameri G, Hu Z, Zhang H, Zhang J, McEachern J, Field H, Daszak P, Eaton BT, Zhang S, Wang LF. Bats are natural reservoirs of SARS-like coronaviruses. Science. 2005;310:676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- 6.Yang L, Wu Z, Ren X, Yang F, He G, Zhang J, Dong J, Sun L, Zhu Y, Du J, Zhang S, Jin Q. Novel SARS-like betacoronaviruses in bats, China, 2011. Emerg Infect Dis. 2013;19:989–991. doi: 10.3201/eid1906.121648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drexler JF, Gloza-Rausch F, Glende J, Corman VM, Muth D, Goettsche M, Seebens A, Niedrig M, Pfefferle S, Yordanov S, Zhelyazkov L, Hermanns U, Vallo P, Lukashev A, Müller MA, Deng H, Herrler G, Drosten C. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J Virol. 2010;84:11336–11349. doi: 10.1128/JVI.00650-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tao Y, Tong S. Complete genome sequence of a severe acute respiratory syndrome-related coronavirus from Kenyan bats. Microbiol Resour Announc. 2019;8 doi: 10.1128/MRA.oo548-19. e00548-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhou H, Ji J, Chen X, Bi Y, Li J, Wang Q, Hu T, Song H, Zhao R, Chen Y, Cui M, Zhang Y, Hughes AC, Holmes EC, Shi W. Identification of novel bat coronaviruses sheds light on the evolutionary origins of SARS-CoV-2 and related viruses. Cell. 2021;184:4380–4391. doi: 10.1016/j.cell.2021.06.008. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murakami S, Kitamura T, Suzuki J, Sato R, Aoi T, Fujii M, Matsugo H, Kamiki H, Ishida H, Takenaka-Uema A, Shimojima M, Horimoto T. Detection and characterization of bat sarbecovirus phylogenetically related to SARS-CoV-2, Japan. Emerg Infect Dis. 2020;26:3025–3029. doi: 10.3201/eid2612.203386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Delaune D, Hul V, Karlsson EA, Hassanin A, Ou TP, Baidaliuk A, Gámbaro F, Prot M, Tu VT, Chea S, Keatts L, Mazet J, Johnson CK, Buchy P, Dussart P, Goldstein T, Simon-Lorière E, Duong V. A novel SARS-CoV-2 related coronavirus in bats from Cambodia. Nat Commun. 2021;12:6563. doi: 10.1038/s41467-021-26809-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wacharapluesadee S, Tan CW, Maneeorn P, Duengkae P, Zhu F, Joyjinda Y, Kaewpom T, Chia WN, Ampoot W, Lim BL, Worachotsueptrakun K, Chen VC, Sirichan N, Ruchisrisarod C, Rodpan A, Noradechanon K, Phaichana T, Jantarat N, Thongnumchaima B, Tu C, Crameri G, Stokes MM, Hemachudha T, Wang LF. Evidence for SARS-CoV-2 related coronaviruses circulating in bats and pangolins in Southeast Asia. Nat Commun. 2021;12:972. doi: 10.1038/s41467-021-21240-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Temmam S, Vongphayloth K, Baquero E, Munier S, Bonomi M, Regnault B, Douangboubpha B, Karami Y, Chrétien D, Sanamxay D, Xayaphet V, Paphaphanh P, Lacoste V, Somlor S, Lakeomany K, Phommavanh N, Pérot P, Dehan O, Amara F, Donati F, Bigot T, Nilges M, Rey FA, Van der Werf S, Brey PT, Eloit M. Bat coronaviruses related to SARS-CoV-2 and infectious for human cells. Nature. 2022;604:330–336. doi: 10.1038/s41586-022-04532-4. [DOI] [PubMed] [Google Scholar]
- 14.Wang N, Li SY, Yang XL, Huang HM, Zhang YJ, Guo H, Luo CM, Miller M, Zhu G, Chmura AA, Hagan E, Zhou JH, Zhang YZ, Wang LF, Daszak P, Shi ZL. Serological evidence of bat SARS-related coronavirus infection in humans. China. Virol Sin. 2018;33:104–107. doi: 10.1007/s12250-018-0012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, Karesh W, Lipkin WI, Morse SS, Consortium PREDICT, Mazet JAK, Goldstein T. Global patterns in coronavirus diversity. Virus Evol. 2017;3 doi: 10.1093/ve/vex012. vex012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin XD, Wang W, Hao ZY, Wang ZX, Guo WP, Guan XQ, Wang MR, Wang HW, Zhou RH, Li MH, Tang GP, Wu J, Holmes EC, Zhang YZ. Extensive diversity of coronaviruses in bats from China. Virology. 2017;507:1–10. doi: 10.1016/j.virol.2017.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wacharapluesadee S, Duengkae P, Rodpan A, Kaewpom T, Maneeorn P, Kanchanasaka B, Yingsakmongkon S, Sittidetboripat N, Chareesaen C, Khlangsap N, Pidthong A, Leadprathom K, Ghai S, Epstein JH, Daszak P, Olival KJ, Blair PJ, Callahan MV, Hemachudha T. Diversity of coronavirus in bats from Eastern Thailand. Virol J. 2015;12:57. doi: 10.1186/s12985-015-0289-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lacroix A, Duong V, Hul V, San S, Davun H, Omaliss K, Chea S, Hassanin A, Theppangna W, Silithammavong S, Khammavong K, Singhalath S, Greatorex Z, Fine AE, Goldstein T, Olson S, Joly DO, Keatts L, Dussart P, Afelt A, Frutos R, Buchy P. Genetic diversity of coronaviruses in bats in Lao PDR and Cambodia. Infect Genet Evol. 2017;48:10–18. doi: 10.1016/j.meegid.2016.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Valitutto MT, Aung O, Tun KYN, Vodzak ME, Zimmerman D, Yu JH, Win YT, Maw MT, Thein WZ, Win HH, Dhanota J, Ontiveros V, Smith B, Tremeau-Brevard A, Goldstein T, Johnson CK, Murray S, Mazet J. Detection of novel coronaviruses in bats in Myanmar. PLoS One. 2020;15 doi: 10.1371/journal.pone.0230802. e0230802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen T, Murray KA, Zambrana-Torrelio C, Morse SS, Rondinini C, Di Marco M, Breit N, Olival KJ, Daszak P. Global hotspots and correlates of emerging zoonotic diseases. Nat Commun. 2017;8:1124. doi: 10.1038/s41467-017-00923-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coker RJ, Hunter BM, Rudge JW, Liverani M, Hanvoravongchai P. Emerging infectious diseases in Southeast Asia: regional challenges to control. Lancet. 2011;377:599–609. doi: 10.1016/S0140-6736(10)62004-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan CW, Chia WN, Qin X, Liu P, Chen MI, Tiu C, Hu Z, Chen VC, Young BE, Sia WR, Tan YJ, Foo R, Yi Y, Lye DC, Anderson DE, Wang LF. A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2-spike protein-protein interaction. Nat Biotechnol. 2020;38:1073–1078. doi: 10.1038/s41587-020-0631-z. [DOI] [PubMed] [Google Scholar]
- 23.Wang LF, Tan CW, Chia WN, Zhu F, Young B, Chantasrisawad N, et al. Differential escape of neutralizing antibodies by SARS-CoV-2 Omicron and pre-emergent sarbecoviruses. Reseacrh Square. 2022 https://www.researchsquare.com/article/rs-1362541/v1 [accessed 25 February 2023] [Google Scholar]
- 24.Tan CW, Chia WN, Young BE, Zhu F, Lim BL, Sia WR, Thein TL, Chen MI, Leo YS, Lye DC, Wang LF. Pan-sarbecovirus neutralizing antibodies in BNT162b2-immunized SARS-CoV-1 survivors. N Engl J Med. 2021;385:1401–1406. doi: 10.1056/NEJMoa2108453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quan PL, Firth C, Street C, Henriquez JA, Petrosov A, Tashmukhamedova A, Hutchison SK, Egholm M, Osinubi MO, Niezgoda M, Ogunkoya AB, Briese T, Rupprecht CE, Lipkin WI. Identification of a severe acute respiratory syndrome coronavirus-like virus in a Leaf-nosed bat in Nigeria. mBio. 2010;1 doi: 10.1128/mBio.00208-10. e00208–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe S, Masangkay JS, Nagata N, Morikawa S, Mizutani T, Fukushi S, Alviola P, Omatsu T, Ueda N, Iha K, Taniguchi S, Fujii H, Tsuda S, Endoh M, Kato K, Tohya Y, Kyuwa S, Yoshikawa Y, Akashi H. Bat coronaviruses and experimental infection of bats, the Philippines. Emerg Infect Dis. 2010;16:1217–1223. doi: 10.3201/eid1608.100208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brisbois B, Rschny J, Fyfe T, Harder H, Parkes M, Allison S, Buse C, Fumerton R, Oke B. Mapping research on resource extraction and health: a scoping review. Extr Ind Soc. 2019;6:250–259. doi: 10.1016/j.exis.2018.10.017. [DOI] [Google Scholar]
- 28.Evans TS, Myat TW, Hom NS, Ricks K, Aung P, Oo ZM, Maw MT, Toe AT, Aung TH, Kuehnert P, Thant KZ, Win YT, Thein WZ, Schoepp R, Johnson CK, Thu HM. Seroepidemiologic survey of Crimean-Congo hemorrhagic fever virus in Myanmar logging communities. Emerg Infect Dis. 2020;27:1709–1713. doi: 10.3201/eid2706.203223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Evans TS, Myat TW, Aung P, Oo ZM, Maw MT, Toe AT, Aung TH, Hom NS, Shein KT, Thant KZ, Win YT, Thein WZ, Gilardi K, Thu HM, Johnson CK. Bushmeat hunting and trade in Myanmar's central teak forests: threats to biodiversity and human livelihoods. Glob Ecol Conserv. 2020;22 doi: 10.1016/j.gecco.2019.e00889. e00889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sánchez CA, Li H, Phelps KL, Zambrana-Torrelio C, Wang LF, Zhou P, Shi ZL, Olival KJ, Daszak P. A strategy to assess spillover risk of bat SARS-related coronaviruses in Southeast Asia. Nat Commun. 2022;13:4380. doi: 10.1038/s41467-022-31860-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson CK, Hitchens PL, Pandit PS, Rushmore J, Evans TS, Young CCW, Doyle MM. Global shifts in mammalian population trends reveal key predictors of virus spillover risk. Proc Biol Sci. 2020;287 doi: 10.1098/rspb.2019.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu K, Pan X, Li L, Yu F, Zheng A, Du P, Han P, Meng Y, Zhang Y, Wu L, Chen Q, Song C, Jia Y, Niu S, Lu D, Qiao C, Chen Z, Ma D, Ma X, Tan S, Zhao X, Qi J, Gao GF, Wang Q. Binding and molecular basis of the bat coronavirus RaTG13 virus to ACE2 in humans and other species. Cell. 2021;184:3438–3451. doi: 10.1016/j.cell.2021.05.031. e10. [DOI] [PMC free article] [PubMed] [Google Scholar]