Abstract
Metastatic tumor is initiated by metastatic seeds (cancer stem cells “CSCs”) in a controlled redox microenvironment. Hence, an effective therapy that disrupts redox balance with eliminating CSCs is critical. Diethyldithiocarbamate (DE) is potent inhibitor of radical detoxifying enzyme (aldehyde dehydrogenase “ALDH”1A) causing effective eradication of CSCs. This DE effect was augmented and more selective by its nanoformulating with green synthesized copper oxide (Cu4O3) nanoparticles (NPs) and zinc oxide NPs, forming novel nanocomplexes of CD NPs and ZD NPs, respectively. These nanocomplexes exhibited the highest apoptotic, anti-migration, and ALDH1A inhibition potentials in M.D. Anderson-metastatic breast (MDA-MB) 231 cells. Importantly, these nanocomplexes revealed more selective oxidant activity than fluorouracil by elevating reactive oxygen species with depleting glutathione in only tumor tissues (mammary and liver) using mammary tumor liver metastasis animal model. Due to higher tumoral uptake and stronger oxidant activity of CD NPs than ZD NPs, CD NPs had more potential to induce apoptosis, suppress hypoxia-inducing factor gene, and eliminate CD44+CSCs with downregulating their stemness, chemoresistance, and metastatic genes and diminishing hepatic tumor marker (α-fetoprotein). These potentials interpreted the highest tumor size reduction with complete eradicating tumor metastasis to liver in CD NPs. Consequently, CD nanocomplex revealed the highest therapeutic potential representing a safe and promising nanomedicine against the metastatic stage of breast cancer.
Subject terms: Breast cancer, Cancer therapy, Pharmaceutics
Introduction
Breast cancer is the second leading cause of cancer death in women globally. Breast cancer mortality is attributed to metastatic growth, not the primary tumor1,2. Liver is the most common and first metastasis target organ of breast tumor3. The breast cancer liver metastasis is triggered by multiple factors in breast cancer stem cell (seed) and liver microenvironment (soil) under a tightly controlled redox balance via lowering cellular reactive oxygen species (ROS) and elevating antioxidant mediators (e.g., glutathione)4,5. One of the key factors in breast cancer cell metastasis (invasion and adhesion) is CD44, which is expressed on cancer stem cells (CSCs)5,6. These CSCs have a high expression of aldehyde dehydrogenase (ALDH) 1-maintained non-ntoxic reactive radical levels to mediate cell invasion, and apoptosis resistance5,7. The elevated ALDH1 level in breast cancer patients is associated with chemoresistance and metastasis. Hence, ALDH1 activates stemness pathways (Notch and Wnt/β-catenin) and hypoxia-inducible factor (HIF)1α/vascular endothelial growth factor (VEGF) pathway8,9. The main factor which modulates the microenvironment of metastatic CSCs is HIF1α by reducing the mitochondrial oxidative stress and enhancing the antioxidant synthesis, thereby promoting chemoresistance10,11. Also, HIF and its related genes (e.g., vascular endothelial growth factor, VEGF and TWIST) maintain angiogenesis and the activated expression of matrix metalloproteases (MMP), E-cadherin, and β-catenin5,12. The latter gene enhances the expression of anti-apoptotic, chemoresistance, MMP genes, and CD44 as well as activates Notch signaling-mediated upregulating stemness genes, which also contribute to chemoresistance6,13,14.
The short median survival rate among patients with metastatic breast cancer is due to the insensitivity to current endocrine therapy, chemotherapy, and immunotherapy3. Therefore, more research is needed to establish an effective treatment for metastatic breast cancer. Yao et al. found a correlation between high ALDH1 expression and the progression of invasive ductal carcinoma in breast cancer patients15. Furthermore, Croker et al. evidenced that knocking out ALDH1A1 inhibited the chemoresistance and metastasis of CD44+ breast cancer cells9. A previous study proved that the anti-metastatic potency of diethyldithiocarbamate (the main metabolite of FDA-approved ALDH1 inhibitor “disulfiram”) in cholangiocarcinoma cell lines via suppressing migration and adhesion16. Moreover, the proapoptotic efficacy of disulfiram (parent of diethyldithiocarbamate), as aldehyde dehydrogenase inhibitor, in collapsing CSCs with suppressing their epithelial-mesenchymal transition (EMT, a crucial step in mediating metastasis), was studied17. Interestingly, the chelating copper-diethyldithiocarbamate complex exhibited a stronger anticancer activity than its parent (disulfiram)18,19. Cvek et al. investigated the anti-breast cancer activity of three types of metal-diethyldithiocarbamate (DE) complexes, including copper(II)-DE, zinc(II)-DE, and nickel-DE. Copper-DE and zinc-DE chelating complexes, but not nickel-DE complex, were found to have powerful proteasome inhibition-dependent anticancer efficacy against in M.D. Anderson-metastatic breast (MDA-MB) 231 cells20. Nevertheless, the preclinical application of DE complex was limited due to its low solubility and less selectivity against cancer cells, which leads to toxicity towards normal cells18. Thus, nanoformulation of metal-DE complex is needed. Ren et al. prepared a nanocomplex of copper-DE, by loading it into organic phase-change nanomaterial comprising of stearic acid and lauric acid as the core and phospholipid as the shell. Then a photochemical agent was added to this nanocomplex to achieve a selective tumor targeting upon photothermal heating against breast cancer. This nanocomplex, which was accumulated selectively in breast tumor with little toxicity in normal tissues, exhibited high anticancer effect and anti-metastatic activity18. Several recent studies illustrated the potent proapoptotic activity of green synthesized copper oxide nanoparticles (NPs) and zinc oxide NPs against breast cancer cell lines21–25. The anticancer activity of such metal oxide NPs is primarily due to their pro-oxidant effects, which induce severe oxidative cellular damage26. Recently, a novel simple nanoformulation of DE containing green chemosynthesized-Cu2O NPs revealed higher oxidant effect-mediated anticancer activity with anti-migration potency than its traditional complex against lung, colon, liver and prostate cancer cells27.
In this current study, for the first time, DE was nanoformulated with green chemically synthesized Cu4O3 NPs or zinc oxide NPs for effective elimination of metastatic breast cancer cells. Hence, the apoptotic and anti-migration activity of these unique nanocomplexes of DE were investigated against the highly metastatic breast cancer cells (MDA-MB 231). Moreover, DE-nanocomplexes were evaluated for their pro-oxidant-mediated apoptotic activity and inhibitory efficacy on key mediators of anti-apoptosis, hypoxia, stemness, chemoresistance, angiogenesis, and metastasis in an orthotopic metastatic breast tumor animal model.
Results
Characterization of the prepared NPs
In this study, DE was nanoformulated with green chemically synthesized CO NPs or ZO NPs. These prepared metal oxide NPs have sizes about 112.5 ± 2.5 nm and 101 ± 4.0 nm, respectively (Supplementary Fig. 1), with zeta potentials about 16.5 ± 1.5 mV and − 12.25 ± 0.75 mV, respectively (Supplementary Fig. 2). The nanoformulations of the produced CD NPs and ZD NPs were affirmed by zetasizer-assessed sizes of 156.5 ± 3.5 nm and 206.5 ± 2.5 nm, respectively (Supplementary Fig. 1), with zeta potentials of − 4.65 ± 0.45 mV and − 14.65 ± 0.65 mV, respectively (Supplementary Fig. 2).
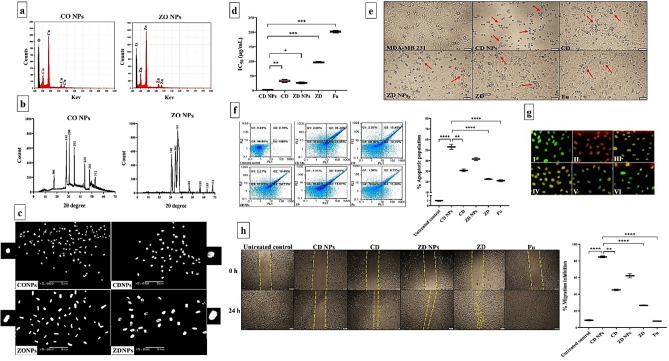
Furthermore, the prepared CO NPs and ZO NPs were characterized using EDX that illustrated the presence of intense signals of Cu (57.69%) and Zn (66.67%), respectively, with oxygen intensities of 42.31% and 33.33%, respectively (Fig. 1a). XRD patterns of CO NPs and ZO NPs correspond to tetragonal Cu4O3 (JCPDS file no. 49-1830) and ZnO (JCPDS file no. 36-1451), respectively (Fig. 1b). Micrographs of SEM (Fig. 1c) show a quasi-spherical structure for CO NPs and its nanocomplex of CD NPs, while ZO NPs and ZD NPs have a rod shape.
Figure 1.
Characterization of the prepared NPs and anticancer impact on MDA-MB 231 cells. (a) Elemental charts of Cu and O in copper oxide (CO) NPs as well as Zn and O in zinc oxide (ZO) NPs using energy dispersive X-ray analysis (EDX). (b) X-ray diffractometer (XRD) patterns for CO NPs and ZO NPs. (c) scanning electron microscope images of CO NPs and ZO NPs as well as their corresponding nanocomplexes with diethyldithiocarbamate (CD NPs and ZD NPs, respectively) at magnification, × 5000; scale bar, 5 µm. For more declaration, individual nanoparticle of each prepared type was showed at maximum magnification. (d) MTT results in the term of IC50 values of CD NPs, CD, ZD NPs, ZD, and 5-fluorouracil (Fu) with (e) morphological alterations of these treated MDA-MB 231 cells (Red arrows refer to apoptotic cells) compared to the untreated control cells. (Magnification, × 100; scale bar, 50 µm). (f) Apoptotic activity as illustrated by flow cytometry dot plots of the untreated MDA-MB 231 and CD NPs-, CD-, ZD NPs-, ZD-, and Fu-treated cells with the total percentage of apoptosis after staining with annexin V/propidium iodide as well as (g) fluorescence microscopic images for the untreated and treated MDA-MB 231 after incubation with acridine orange and ethidium bromide (Green, yellow and orange-red fluorescence reflect healthy, early apoptotic, and late apoptotic cell population, respectively) (Magnification, × 100; scale bar, 50 µm). (h) Anti-migration activity as showed by microscopic pictures of the wound area (yellow dashed line-labeled) in the untreated and treated MDA-MB 231 cells at 0 and 24 h (Magnification, × 40; scale bar, 200 µm) with the percentage of migration inhibition. Data are demonstrated as mean ± SEM. CD NPs were compared to other tested compounds, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***.
In vitro anticancer activity
Cytotoxic, proapoptotic, and anti-migratory impact against MDA-MB 231 cells
Cytotoxicity of the prepared NPs (CO NPs, ZO NPs, CD NPs, and ZD NPs) was determined, in the term of IC50, compared to CD, ZD, DE, copper chloride, zinc chloride, and standard chemotherapy (fluorouracil “Fu”) against MDA-MB 231 cells (Fig. 1d). Based on the lowest IC50 indicating the highest anticancer activity (Fig. 1d), nanocomplexes of CD NPs and ZD NPs exhibited the strongest growth inhibitory effect against MDA-MB 231 cells compared to their corresponding traditional complexes (160.78 ± 3.55 μg/mL and 173.99 ± 5.14 μg/mL, respectively), Fu (184.19 ± 3.93 μg/mL), and DE (201.48 ± 1.87 μg/mL). The IC50 values of copper chloride and zinc chloride could not be calculated due to their maximum utilized concentration did not reach 50% death in the treated MDA-MB 231. Importantly, IC50 of CD NPs (31.93 ± 5.14 μg/mL) was significantly lower than that of ZD NPs (95.98 ± 1.69 μg/mL). Figure 1e illustrates the highest drastic alterations in the morphology of CD NPs-treated MDA-MB 231 cells compared to CD-, ZD NPs-, ZD-, and Fu-treated cells.
Flow cytometry analysis (Fig. 1f) of the apoptotic population (%) for the treated MDA-MB 231 cells clarified that both nanocomplexes (> 40%) were the highest compared to CD (30.81%), ZD (22.58%), and Fu (21.04%). As shown in Fig. 1f, the proapoptotic effect of CD NPs (55.30 ± 4.75%) was significantly higher than that of ZD NPs (41.60 ± 1.44%). These results were confirmed by fluorescence microscope after staining with EB/AO, where green, yellow and reddish-orange fluorescence nuclei reflect healthy, early apoptotic, and late apoptotic cells, respectively (Fig. 1g). Subsequently, the late apoptotic population was only observed in CD NPs-treated MDA-MB 231 cells, whereas the other treated cells refer to the early apoptosis stage (Fig. 1f,g).
Regarding wound healing assay (Fig. 1h), nanocomplexes and traditional complexes revealed a 40-fold higher anti-migratory effect than Fu. Moreover, both nanocomplexes demonstrated higher anti-migration potency than their corresponding conventional complexes in the treated MDA-MB 231 cells. CD NPs inhibited cell migration by 84.83 ± 1.09%, whereas ZD NPs suppressed migration by 62.27 ± 2.59% (Fig. 1h).
Inhibitory potential on ALDH1 activity
Figure 2a demonstrates that CD NPs had the strongest inhibitory ALDH1A potential (83.40%) compared to other prepared DE complexes and Fu. Notably, CDNPs were more effective than ZD NPs (69.86%) for suppressing ALDH1A activity. On the other hand, Fu exhibited the lowest inhibition potency (< 4%) for ALDH1A.
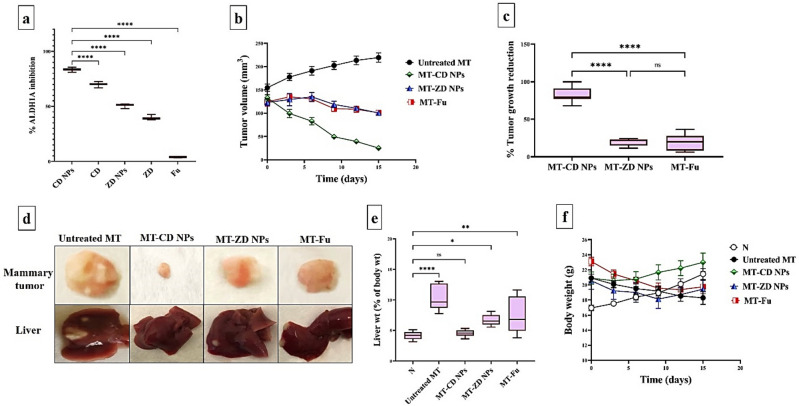
Figure 2.
In vitro ALDH1A inhibitory potential and inhibitory impact on tumor growth using orthotopic mammary tumor animal model. (a) The relative ALDH1A inhibition in the treated MDA-MB 231 cells. (b) The recorded mammary tumor volume using caliper with (c) the reduction in mammary tumor weight that was measured, at the end of experiment, in all three treated groups compared to the untreated mammary tumor-bearing mouse group (MT). Data are demonstrated as mean ± SEM. MT-CD NPs group was compared to other groups, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***. (d) Macroscopic images of mammary tumors and liver tumor nodules of the untreated and treated MT groups. (e) Liver weight, relative to body weight, for untreated and treated MT groups compared to the healthy control group (N). (f) Body weight for untreated and treated MT groups compared to N group. All values are demonstrated as mean ± SEM. Healthy (N) untreated group was compared to other groups, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***.
Therapeutic potency against metastatic mammary tumor (in vivo)
Inhibitory impact on tumor volume and weight
The anti-tumor efficacy of the most active compounds (CD NPs and ZD NPs), in comparison with Fu, was studied in vivo using an orthotopic mammary tumor (MT) model. During 2 weeks of treatment, six injections of CD NPs were able to decrease the tumor size mean, from 118.74 to 25.11 mm3, significantly more than ZD NPs and Fu. In contrast to CD NPs-treated MT group (MT-CD NPs), ZD NPs-treated MT group (MT-CD NPs), and Fu-treated MT group (MT-Fu NPs), the tumor size of the untreated MT group increased from 113.20 to 219.38 mm3 during 2 weeks (Fig. 2b). The tumor weighing revealed that MT-CD NPs group had the highest reduction (81.11 ± 4.07%) in tumor growth, compared to 18.42 ± 2.44% and 18.99 ± 4.11% for MT-ZD NPs and MT-Fu groups, respectively (Fig. 2c). Additionally, macroscopic images of mammary tumors (Fig. 2d) support the superior inhibitory potential of CD NPs on mammary tumor growth and metastasis, as evidenced by the disappearance of liver tumor nodules, compared to ZD NPs and Fu as well as the untreated MT. Furthermore, the untreated MT group had the largest weights of liver organ relative to its corresponding body weight (7.724 ± 1.69%) compared to all treated MT groups and N group (Fig. 2e). This figure also shows no significant difference between liver index of N group and MT-CD NPs group was recorded, but there is statistical variation with MT-ZD NPs and MT-Fu groups. According to mice body weights, the highest reduction in body weight was observed in the untreated MT group compared to all treated groups and N group. Particularly, body weight of MT-CD NPs group was stable as compared to healthy N group (Fig. 2f).
Histological analysis of the therapeutic efficacy of nanocomplexes
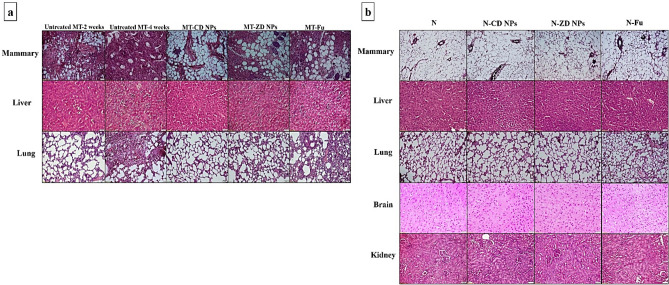
After 2 weeks of orthotopic injection of Ehrlich cells, the incidence of a metastatic mammary tumor was confirmed by H&E staining mammary, liver, and lung tissues that illustrated the mixed ductal and lobular mammary carcinoma, hepatic tumor cells, and normal lung, respectively (Fig. 3a). Following 2 weeks of treatment with nanocomplexes or Fu, H&E staining declared that CD NPs had the best therapeutic potential of CD NPs on mammary tumor with complete suppression of metastasis to liver, compared to ZD NPs and Fu. Unlike the untreated MT and MT-Fu, both nanocomplexes completely halted lung hyperplasia (Fig. 3a). Moreover, H&E stained mammary, liver, lung, brain, and kidney tissues of N-CD NPs, N-ZD NPs, and N-Fu groups revealed no histological alterations when compared to N group (Fig. 3b).
Figure 3.
Histological analysis of anti-tumor potential and toxicity on normal tissues. (a) Hematoxylin and eosin (H&E)-stained tissue sections of the untreated mammary tumor-bearing mouse group (MT) after 2 weeks and 4 weeks of Ehrlich cell orthotopic injection (showing the mixed ductal and lobular mammary carcinoma with invasion to liver and lung hyperplasia) as well as the treated MT groups. (b) H&E-stained tissue sections of healthy untreated group (N) and treated N groups (Magnification, × 200; scale bar, 50 µm).
Selective accumulation of nanocomplexes in tumor tissues
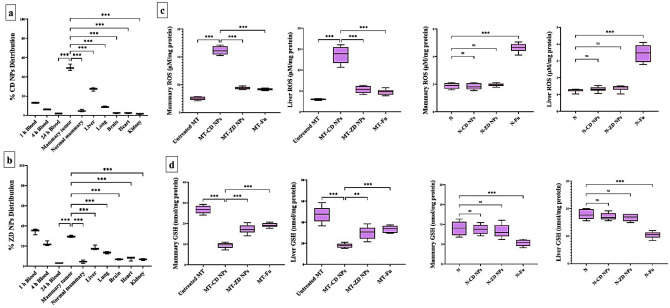
The biodistribution of nanocomplexes was studied, separately, by i.v. injection of CD NPs and ZD NPs into MT mice, after 4 weeks of Ehrlich cell suspension injection. The results of atomic absorption spectroscopy demonstrated that 13.00% and 6.187% of CD NPs and 34.48% and 22.34% of ZD NPs remained in blood at 1 h and 4 h post injections, respectively, in the treated MT mice (Fig. 4a,b). After 24 h, CD NPs and ZD NPs distributed mostly (49.71% and 32.25%, respectively) in mammary tumor, followed by liver (27.10% and 18.22%, respectively) and lung (8.930% and 13.48%, respectively). The other investigated tissues (brain, heart and kidney) and blood contained the lowest amounts of CD NPs (< 2.5%) and ZD NPs (< 7%) as shown in Fig. 4a,b.
Figure 4.
Nanocomplexes’ selective biodistribution in tumor tissues and selective ROS elevation with GSH depletion impacts. Selective biodistribution of (a) CD NPs and (b) ZD NPs in blood and tissues. All values are demonstrated as mean ± SEM. Mammary tumor was compared to other tissues, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***. (c) ROS level (μM/mg protein) and (d) GSH content (nmol/mg protein) in mammary tumor and liver of the untreated mammary tumor-bearing mouse group (MT), CD NPs-treated MT group (MT-CD NPs), MT-ZD NPs group, and MT-Fu group compared to normal mammary and liver tissues of healthy control (N) untreated group, CD NPs-treated N group (N-CD NPs), N-ZD NPs, and N-Fu. All values are demonstrated as mean ± SEM. MT-CD NPs group was compared to other groups, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***.
Selective pro-oxidant effect of nanocomplexes
In mammary tissues of the treated MT groups, CD NPs, ZD NPs, and Fu enhanced ROS generation by 4.431, 1.736, and 1.656 folds, respectively, relative to the untreated MT. Also, MT-CD NPs, MT-ZD NPs, and MT-Fu groups showed an increase in hepatic ROS levels by 2.708-, 1.060-, and 1.000-folds, respectively, compared to the untreated MT (Fig. 4c). This ROS elevation was associated with GSH depletion, CD NPs (~ threefold) outperforming ZD NPs (~ 1.5-fold) and Fu (~ 1.4-fold) in diminishing GSH level in both mammary tumor and liver tissues relative to the untreated MT (Fig. 4d). In contrast to Fu, both nanocomplexes did not cause any significant elevation of ROS or reduction in GSH levels in the normal mammary and liver tissues of N-CD NPs and N-ZD NPs groups relative to N group (Fig. 4c,d).
Immunohistochemical investigation of the therapeutic efficacy of nanocomplexes
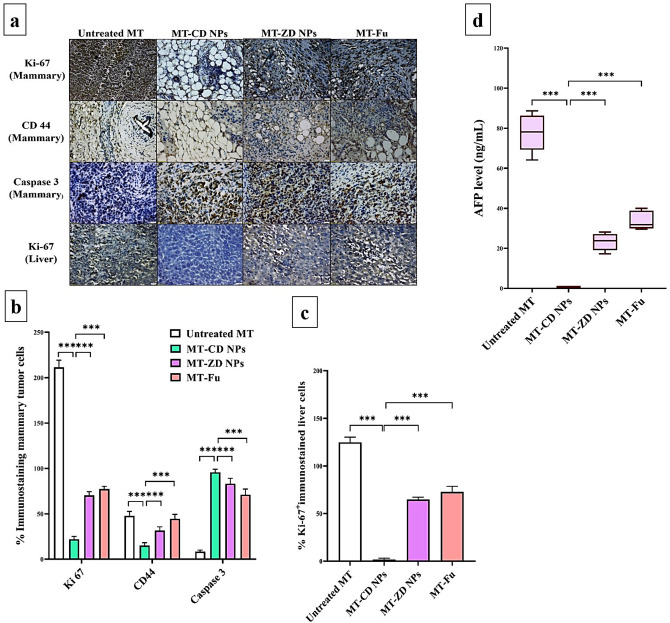
The immunostaining tumor tissues of mammary and liver with Ki-67 (Fig. 5a–c) illustrated that CD NPs exhibited the highest suppressive potential on this proliferative marker, by 9.61 and 69.45-folds, respectively, compared to ZD NPs (3.00 and 1.92-folds, respectively) and Fu (2.73 and 1.71-folds, respectively). Also, CD NPs demonstrated the highest anti-CSC and proapoptotic activities in mammary tumor, as evidenced by the minimum percentage of CD44+-stained cells (20.26 ± 2.19%) and the maximum percentage of caspase 3+-stained cells (95.71 ± 1.49%) compared to ZD NPs and Fu. There is no significant difference was recorded between Zn NPs and Fu in the terms of % Ki-67+-and caspase 3+-immunostained tumors. However, the percentage of CD44+-stained mammary tumor cells of MT-Zn NPs group was significantly lower than that of MT-Fu group, p < 0.05 (Fig. 5a–c).
Figure 5.
Immunohistochemical analysis and biochemical detection of anti-metastatic tumor potential. (a) Microscopic images of Ki-67, CD44, and caspase 3-immunostained mammary tumor as well as Ki-67-immunostained liver sections of the untreated mammary tumor-bearing mouse group (MT), CD NPs-treated MT group (MT-CD NPs), MT-ZD NPs group, and MT-Fu group with (b,c) their percentages of positive immunostained tumor cells. (d) The blood level (ng/mL) of α-fetoprotein (AFP). All values are demonstrated as mean ± SEM. MT-CD NPs group was compared to other groups, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***.
Suppressive effect of nanocomplexes on liver tumor marker
The main liver tumor marker (α-fetoprotein) which was used to evaluate the anti-metastatic potential of the prepared nanocomplexes, was elevated abnormally in the untreated MT group by > 77 folds relative to the healthy N group. As shown in Fig. 5d, CD NPs can normalize α fetoprotein (AFP < 0.6 ng/mL), whereas its level was still high in MT-ZD NPs group (23.25 ± 1.94 ng/mL) and MT-Fu group (33.89 ± 2.08 ng/mL).
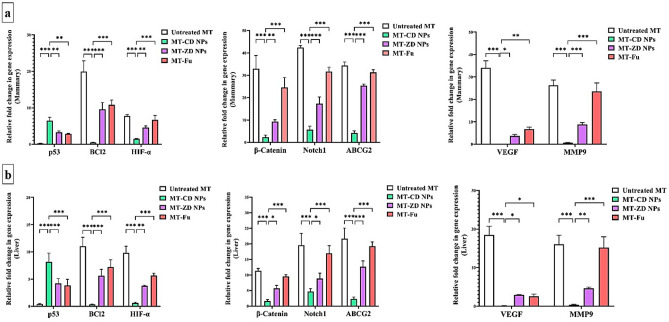
Regulatory effect of nanocomplexes on the expression of the main proapoptotic gene and oncogenes
Figure 6a,b shows that CD NPs exhibited the highest activity for upregulating the expression of the main proapoptotic gene (p53), by 6.461 and 8.141-folds compared to ZD NPs (3.272 and 4.213-folds) and Fu (2.858 and 3.857-folds) in mammary tumor and liver tissues, respectively. There is no significant variation in the ability of ZD NPs and Fu to induce p53 expression. Furthermore, CD NPs had the highest potential to repress key anti-apoptotic gene (BCl2), HIF-α-driven hypoxic microenvironment, β-catenin- and Notch1-mediated stemness, ABCG2-dependent chemoresistance, VEGF-dependent angiogenesis, and MMP9-stimulated metastasis, in both mammary and liver tumor tissues. ZD NPs suppressed the above-mentioned oncogenes more effectively than Fu, excluding HIF, VEGF, and BCl2 expression which showed a comparable inhibitory effect between ZD NPs and Fu (Fig. 6a,b).
Figure 6.
Regulatory impact on the expression of key apoptotic gene and oncogenes using qPCR. The relative gene expression of p53, BCl2, hypoxia-inducing factor (HIF)-α, β-catenin, Notch1, ATP Binding Cassette Subfamily G Member 2 (ABCG2), vascular endothelial growth factor (VEGF), and matrix metalloprotease (MMP)9 were quantified in (a) mammary tumor and (b) liver tissues of the untreated mammary tumor-bearing mouse group (MT), CD NPs-treated MT group (MT-CD NPs), MT-ZD NPs group, and MT-Fu group. All values are demonstrated as mean ± SEM. MT-CD NPs group was compared to other groups, considering significantly different at p < 0.05*, < 0.01**, and < 0.001***.
Safety of these nanocomplexes on liver, kidney and hematological parameters
For investigating the in vivo toxicity profile of these nanocomplexes, functional parameters of liver (ALT, AST, and albumin) and kidney (urea and creatinine) as well as CBC were assessed in N and MT groups that were treated with CD NPs and ZD NPs versus the untreated groups (Tables 1, 2). Table 1 reveals that both nanocomplexes did not cause any significant variation in the functional parameters of liver (ALT, AST, and albumin) or kidney (urea and creatinine) in either the treated N or treated MT groups, relative to the healthy N group. Meanwhile, Fu caused a substantial increase in liver function parameters, including ALT and AST, in N-Fu and MT-Fu groups. On the other hand, the untreated MT group demonstrated significant abnormal alterations in liver parameters, including elevation of ALT and AST with diminishing albumin levels, and a moderate increase in kidney functions, compared to healthy N group (Table 1).
Table 1.
Liver and kidney functional parameters.
| N | N-CD NPs | N-ZD NPs | N-Fu | MT | MT-CD NPs | MT-ZD NPs | MT-Fu | |
|---|---|---|---|---|---|---|---|---|
| ALT (U/L) | 99.21 ± 3.15 | 96.99 ± 2.74 | 92.02 ± 0.63 | 108.4 ± 6.65 | 138.4 ± 4.68*** | 93.69 ± 3.07 | 110.1 ± 4.16 | 119.0 ± 2.03** |
| AST (U/L) | 186.1 ± 5.87 | 177.0 ± 9.95 | 184.2 ± 7.61 | 208.3 ± 13.8 | 203.7 ± 4.33** | 184.9 ± 7.48 | 199.2 ± 3.49 | 205.8 ± 3.47** |
| Albumin (g/dL) | 3.989 ± 0.31 | 4.305 ± 0.26 | 4.166 ± 0.29 | 4.267 ± 0.26 | 3.040 ± 0.12** | 4.323 ± 0.20 | 3.483 ± 0.06 | 3.360 ± 0.11* |
| Urea (mg/dL) | 47.64 ± 3.04 | 48.25 ± 1.45 | 49.13 ± 1.01 | 49.38 ± 0.83 | 56.33 ± 4.65 | 44.65 ± 5.21 | 46.54 ± 2.68 | 49.62 ± 2.77 |
| Creatinine (mg/dL) | 1.844 ± 0.08 | 1.707 ± 0.19 | 1.644 ± 0.15 | 1.679 ± 0.14 | 1.969 ± 0.05 | 1.608 ± 0.12 | 1.757 ± 0.15 | 1.678 ± 0.16 |
N the untreated healthy control normal mouse group, N-CD NPs nanocomplex of Cu4O3 with diethyldithiocarbamate (DE)-treated normal mouse group, N-ZD NPs nanocomplex of ZnO with DE-treated normal mouse group, N-Fu 5-fluorouracil-treated normal mouse group, MT the untreated mammary tumor-bearing mouse group, MT-CD NPs CD NPs-treated MT group, MT-ZD NPs ZD NPs-treated MT, and MT-Fu Fu-treated MT group.
All values are demonstrated as mean ± SEM. N group was compared to other groups and considered significantly different at p < 0.05*, < 0.01**, and < 0.0001***.
Table 2.
Hematological parameters.
| N | N-CD NPs | N-ZD NPs | N-Fu | MT | MT-DC NPs | MT-DZ NPs | MT-Fu | |
|---|---|---|---|---|---|---|---|---|
| RBC | ||||||||
| RBC (106/µL) | 7.99 ± 0.09 | 8.13 ± 0.13 | 7.53 ± 0.47 | 7.10 ± 0.10 | 6.84 ± 0.16 | 8.04 ± 0.33 | 7.35 ± 0.34 | 6.33 ± 0.43 |
| Hg (g/dL) | 13.7 ± 0.70 | 14.2 ± 0.11 | 12.6 ± 0.40 | 13.3 ± 0.22 | 9.85 ± 0.75* | 11.1 ± 0.35 | 10.8 ± 0.90 | 9.40 ± 1.20* |
| HCT (%) | 35.4 ± 1.09 | 35.7 ± 0.29 | 33.1 ± 1.85 | 33.5 ± 0.48 | 26.2 ± 1.60* | 29.2 ± 1.45 | 27.9 ± 2.20 | 25.3 ± 2.65* |
| MCV (fL) | 34.6 ± 0.65 | 34.8 ± 0.37 | 34.6 ± 0.37 | 36.7 ± 0.45 | 38.2 ± 1.35 | 36.3 ± 0.25 | 37.8 ± 1.25 | 40.0 ± 1.45* |
| MCH (pg) | 17.4 ± 0.40 | 17.4 ± 0.12 | 17.7 ± 0.40 | 18.4 ± 0.30 | 14.3 ± 0.75 | 16.8 ± 0.15 | 14.6 ± 0.55 | 14.8 ± 0.90 |
| MCHC (g/dL) | 39.1 ± 0.15 | 39.8 ± 0.15 | 39.4 ± 0.28 | 39.0 ± 0.09 | 37.5 ± 0.50 | 38.0 ± 0.65 | 38.6 ± 0.20 | 37.0 ± 1.00 |
| RDW-CV (%) | 13.2 ± 0.2 | 13.8 ± 0.15 | 15.3 ± 0.2 | 15.0 ± 0.25 | 21.9 ± 0.80* | 16.4 ± 1.2 | 21.8 ± 3.15 | 20.2 ± 0.30* |
| RDW-SD fL) | 21.4 ± 0.40 | 21.2 ± 0.25 | 24.2 ± 0.15 | 25.6 ± 0.51 | 30.2 ± 0.05* | 28.6 ± 2.05 | 29.7 ± 3.40 | 29.1 ± 1.45* |
| WBC | ||||||||
| WBC (103/µL) | 9.11 ± 0.21 | 10.5 ± 0.55 | 7.95 ± 0.55 | 7.30 ± 0.40 | 7.90 ± 0.40 | 10.6 ± 0.90 | 12.2 ± 0.2 | 13.8 ± 1.05* |
| Lymph% | 83.4 ± 0.10 | 83.7 ± 0.70 | 80.6 ± 0.40 | 85.3 ± 0.35 | 54.8 ± 1.50 | 83.8 ± 0.45*** | 75.6 ± 6.5*** | 47.3 ± 4.70*** |
| Mid% | 6.20 ± 0.20 | 5.05 ± 0.55 | 8.70 ± 0.30 | 5.50 ± 0.40 | 15.3 ± 2.20 | 5.25 ± 0.15 | 11.2 ± 0.80* | 19.6 ± 0.10 |
| Gran% | 10.4 ± 0.10 | 11.2 ± 1.25 | 10.7 ± 0.70 | 9.15 ± 0.75 | 29.9 ± 0.70*** | 10.9 ± 0.60*** | 18.55 ± 1.95*** | 33.1 ± 4.60*** |
| PLT | ||||||||
| PLT (103/µL) | 874 ± 45.0 | 842 ± 42.0 | 844 ± 5.50 | 816 ± 6.00 | 955 ± 53.0 | 1006 ± 75.0 | 1057 ± 93.0 | 802 ± 21.0 |
| MPV (fL) | 5.66 ± 0.16 | 5.56 ± 0.23 | 5.50 ± 0.30 | 5.98 ± 0.31 | 6.05 ± 0.25 | 5.70 ± 0.10 | 6.05 ± 0.15 | 6.25 ± 0.04 |
| PDW (fL) | 14.8 ± 0.14 | 15.1 ± 0.15 | 15.5 ± 0.5 | 14.7 ± 0.63 | 14.7 ± 0.05 | 14.5 ± 0.03 | 14.6 ± 0.05 | 14.5 ± 0.16 |
| PCT (mL/L) | 4.62 ± 0.08 | 4.08 ± 0.57 | 3.97 ± 0.28 | 4.91 ± 0.24 | 5.81 ± 0.54 | 5.72 ± 0.51 | 6.40 ± 0.71 | 5.04 ± 0.15 |
N the untreated healthy control normal mouse group, N-CD NPs nanocomplex of Cu4O3 with diethyldithiocarbamate (DE)-treated normal mouse group, N-ZD NPs nanocomplex of ZnO with DE-treated normal mouse group, N-Fu 5-fluorouracil-treated normal mouse group, MT the untreated mammary tumor-bearing mouse group, MT-CD NPs CD NPs-treated MT group, MT-ZD NPs ZD NPs-treated MT, and MT-Fu Fu-treated MT group, RBC red blood cell, Hg hemoglobin, HCT hematocrit, MCV mean corpuscular volume, fL 10–15 L, MCH mean corpuscular hemoglobin, MCHC mean corpuscular hemoglobin concentration, RDW-CV and RDW-SD red blood cell distribution width-coefficient of variation and standard deviation, WBC white blood cell, Lymph lymphocyte, Mid monocytes, eosinophils and basophils, Gran granulocyte, PLT platelet, MPV mean platelet volume, PDW platelet distribution width, and PCT plateletcrit.
All values are demonstrated as mean ± SEM. N group was compared to other groups and considered significantly different at p < 0.05*, < 0.01**, and < 0.0001***.
Regarding hematological parameters (Table 2), N-CD NPs and MT-CD NPs groups did not show any abnormal changes, relative to the healthy N group, in RBCs count, hemoglobin (Hg), and their related factors, and the total count or differential percentages of WBCs, as well as platelet (PLT) count, volume, distribution width, and plateletcrit (PCT). Both N-ZD NPs and N-Fu groups also did not demonstrate any significant hematological alterations relative to N group. On the other side, MT group revealed a significant decrease in Hg, hematocrit (HCT), and lymphocyte (Lymph) % as well as a statistical increase in red blood cell distribution width-coefficient of variation and standard deviation (RDW-CV and RDW-SD), monocyte (Mid) %, and neutrophil (Gran)% when compared to N group. In MT-Fu NPs, these abnormalities cannot be normalized by Fu which also caused a significant elevation in mean corpuscular volume (MCV) and WBCs count compared to N group. Meanwhile, MT-ZD NPs group showed a slight decrease in Hg, HCT, and Lymph% as well as a little increase in RDW-CV, RDW-SD, and neutrophil (Gran)% with a significant increase in Mid% (Table 2).
Discussion
Worldwide, breast cancer is the most common malignancy-related death in women. The survival rate decreases dramatically, among patients with metastatic tumor, mainly attributed to the poor response to standard therapy1. To the best of our knowledge, this is the first study that nanoformulated DE with Cu4O3 NPs and ZnO NPs to investigate their activities on metastatic breast tumor. The generated CD and ZD nanocomplexes had nanosizes (156.5 nm and 206.5 nm, respectively), negative zeta potential, and morphology similar to their corresponding metal oxide NPs (semi-sphere and rods, respectively). In vitro anticancer assessment revealed that CD NPs was the most potent growth inhibitory activity against metastatic breast cancer cells (MDA-MB 231) with the lowest IC50 (31.93 μg/mL), and the highest proapoptotic and anti-metastatic potentials compared to ZD NPs and Fu. Moreover, CD NPs demonstrated the highest antitumor effect in terms of reducing mammary tumor volume and weight as well as histological, immunohistochemical, biochemical, and molecular analyses of oncogenic and apoptotic markers in mammary tumor and liver tissues. These current in vitro and in vivo demonstrations of the strongest anticancer potency of CD NPs are mostly linked to their pro-oxidant activity, as evidenced by the strongest ALDH1A inhibition in MDA-MB 231 cells and the highest ROS fold increment with GSH depletion in tumor tissues (mammary and liver) (Figs. 2a, 4c,d). Previous studies illustrated the oxidant activity of DE via its high thiol affinity, forming disulfide adducts in enzymatic and nonenzymatic antioxidant (such as ALDH1 and GSH, respectively)28,29. Herein, the anticancer activity-dependent pro-oxidant activity of DE was potentiated by metal oxide NPs and vice versa, as evidenced by lowering IC50 values of both nanocomplexes compared to DE or metal oxide NPs.
The powerful inducible oxidative stress after treatment with CD NPs rather than ZD NPs is primarily attributed to the nature of the contained metal oxide NPs (Cu4O3 and ZnO, respectively), as well as oxygen intensity that was higher in CO NPs than ZO NPs by 8.98% based on EDX analysis. Regarding CO NPs of CD nanocomplex, Cu4O3 is copper (I, II) oxide mineral with formula (Cu+1)2 (Cu+2)2 O3. In contrast to Cu2O’s direct hydroxyl radical generation effect, CuO’s production of hydroxyl radical requires GSH for reducing Cu+2 to Cu+1 which catalyzes Fenton reaction, leading to GSH depletion30,31. The decreased GSH is also related to the direct binding of Cu to its thiol group and the oxidative effect of the generated hydroxyl radicals, resulting in an increased formation of its oxidized form (GSSG) and, subsequently, loss of its function that affects negatively on the total cellular antioxidant capacity31. Cu has a higher binding affinity with greater stability constant for thiol groups than Zn32. Moreover, in consistence with the current findings, a previous comparison study found that CuO caused more oxidative DNA damage with extensive mitochondrial depolarization than ZnO in A549 cells26. The GSH lowering and ROS overproduction effects of these metal oxide NPs caused severe oxidative stress (oxidation of DNA, protein, and lipid) leading to selective cancer cell death with minimal toxicity to normal cells30,33. This selectivity is mainly due to their nanosize as well as abnormal tumor vascular (dilated and disorganized vessels) which enhance the permeability and retention effect of NPs selectively in tumor tissues while halting their penetration into normal vessels of healthy tissues34. As it was demonstrated, both nanocomplexes elevated ROS and lowered GSH only in tumor tissues (mammary and liver) without any significant changes between the treated normal tissues and healthy control untreated tissues. Furthermore, no marked variation in histology analysis and biochemical parameters was observed between N, N-CD NPs and N-ZD NPs groups. Obviously, the biodistribution of both nanocomplexes was recorded mostly (85.74% and 63.95%, respectively) in tumor tissues with only minimal percentages in normal tissues. This also indicates that CD NPs had significantly higher tumor uptake than ZD NPs (p < 0.01), which could be attributed to CD NPs’ smaller size, lesser negative charge, and semi-spherical shape. It is well understood that NPs with smaller sizes, spherical shapes, and slight negative charge achieve better circulation and tumor accumulation34,35.
Accordingly, CD NPs exhibited stronger selective oxidant activity, with higher tumoral uptake, which is the main contributor to inducing selective apoptosis coupled with inhibiting stemness and metastasis, higher than ZD NPs (Figs. 4, 5, 6). Overgeneration of ROS without covering by the cellular antioxidant system causes p53 expression- and activation-dependent DNA damage and mitochondrial membrane potential loss, leading to a downregulation of BCL2 and release of cytochrome C from mitochondria, and ultimately, inducing caspase-dependent apoptosis. Furthermore, GSH lowering causes apoptosis induction due to the loss of S-glutathiolated protection of GSH for caspase 3 from proteolytic cleavage activation36. There is a strong correlation between ROS detoxify system (e.g., ALDH1 and GSH) and CSC metastatic and self-renewal potentials. Low ROS level is critical for CSC maintenance by enhancing PI3K/AKT/mTOR activation of HIF1α-mediated hypoxia microenvironment. HIF is a crucial enhancer of GSH synthesis, glycolytic genes, Notch signaling pathway, and expression of CD44, VEGF, and ABCG2 leading to the stimulation of stemness, angiogenesis, and chemoresistance37,38. The latter is also induced by the overexpression of ALDH1 as well as the promotion of stemness and metastasis pathways (including, Notch1, Wnt/β-catenin, and EMT)39. Previous studies found that CO NPs and disulfiram-Cu complex induced ROS-mediated apoptosis by suppressing AKT signaling pathway40,41. HIF-1α suppression with uncontrolled ROS production leads to the elimination of CSC (metastatic seed) with blocking its stemness and metastasis potentials and restoring its sensitivity to chemotherapy4,6,38. In line with these previous findings6,30, the strongest oxidant impact of CD NPs with potent suppressing HIF-1α was linked to the lowest percentage of CD44+ and Ki-67+ immunostaining and the highest downregulation of stemness, metastasis, and VEGF genes, compared to ZD NPs and Fu (Figs. 5a–c, 6a,b). Thus, oxidant-mediated apoptosis limits tumor transformation and metastasis, as confirmed by histological analysis of the marked elimination of mammary tumor cells with complete inhibition of their metastasis to liver or lung, and immunostaining Ki-67 decline with the lowest MMP9 expression and hepatic tumor marker (AFP) level in MT-CD NPs group, compared to MT-ZD NPs and MT-Fu. Moreover, CD NPs preferentially eradicated metastatic tumor without any alterations in normal values of liver or kidney functional indicators and hematological parameters, as demonstrated in Tables 1 and 2.
In conclusion, these new nanoformulations of DE with green chemically synthesized CO NPs (Cu4O3) and ZO NPs exhibited a selective oxidant potential-dependent anticancer potency, compared to Fu, using metastatic breast cancer cells (MDA-MB 231) and metastatic mammary tumor animal model. Importantly, CD NPs demonstrated higher tumoral uptake and stronger oxidant activity than ZD NPs. This massive oxidative stress in MT-CD NPs was associated with the strongest HIF1α suppression, caspase 3-dependent apoptosis induction, and CD44+CSCs (seeds of metastasis) elimination with blocking their stemness, chemoresistance, and metastasis gene expression compared MT-ZD NPs and MT-Fu. Hence, histological analysis and immunostaining with Ki-67 and caspase 3 as well as lowering MMP-9 expression and liver tumor marker (AFP) level confirmed that CD NPs had the best therapeutic potential for the eradication of metastatic mammary tumor (mixed ductal and lobular mammary carcinoma), compared to ZD NPs and Fu. Besides that, CD NPs did not show any marked variation in the functional parameters of liver and kidney as well as hematological indexes, compared to healthy control group. Collectively, this novel nanocomplex of DE and Cu4O3 NPs was more effective than ZD nanocomplex in annihilating metastatic breast cancer.
Materials and methods
Materials
Chitosan was obtained from Acros Organics (New Jersey, USA). Dulbecco’s modified Eagle’s medium (DMEM) and fetal bovine serum (FBS) were obtained from GIBCO (New York, USA). Standard chemotherapy (fluorouracil) was from Applichem for pharmaceuticals Co. (Darmstadt, Germany). 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), retinal, 2,7 dichlorofluorescence diacetate (DCFDA), nuclear stains, horseradish peroxidase-conjugated secondary antibody, 3,3′ diaminobenzidine, glutathione (GSH) were purchased from Sigma-Aldrich (Burlington, Massachusetts, USA). Trypan Blue, primary antibodies of ki-67, CD 44, and caspase 3, RNA extraction kit, and primers were obtained from Thermo Fisher Scientific (Waltham, Massachusetts, USA). One-step qPCR SYBR green master mix kit and α-Fetoprotein electrochemiluminescence detection kit were from GeneDireX (Taoyuan, Taiwan) and Roche Diagnostics (Basel, Switzerland), respectively.
Methods
Preparation and characterization of NPs
Copper oxide nanoparticles were prepared by mixing copper chloride (source of copper salt) with chitosan (capping agent), and vitamin C (reducing agent) as recently described27. ZO NPs were prepared using the precipitation method42 with some modifications, including zinc chloride (0.2 mL, 160 mM) was mixed with NaOH (10 mL, 1 M) and vitamin C (10 mL, 20%), pH was adjusted to 7, and heated for 1 h at 80 °C. Then ZO NPs were obtained after centrifugation. Zetasizer Nano ZS (Malvern, UK) was used to assess the size and zeta potential. Compositional feature, oxidation state (structural property), and morphology were determined using energy dispersive X-ray analysis (EDX, JEOL JEM-1230, Japan), X-ray diffractometer (XRD, Bruker MeaSrv (D2-208219) fitted with Cu Kα radiation tube (λ = 1.5406 Å)27, and scanning electron microscopes (SEM, JEOL JSM (5300), Japan)27, respectively. The latter instrument's data was analyzed utilizing Joint Committee on Powder Diffraction Standards (JCPDS).
These characterized NPs (0.02 mg/mL) were mixed with DE (0.2 mg/mL) to form nanocomplexes of CD NPs and ZD NPs, respectively. The size, zeta potential, and morphology of these nanocomplexes were then determined as mentioned above. In parallel, traditional complexes of copper chloride-DE (CD) and zinc chloride-DE (ZD) were prepared by the same ratio.
In vitro evaluation of anti-breast cancer activity
Determination of cancer growth inhibition by MTT assay
Human breast cancer cell line (MDA-MB 231) which was obtained from American Type Culture Collection (ATCC), seeded (5 × 103 cells, passage no. 41) in 96-well cell culture plates using DMEM medium with 10% FBS as the culture medium, and incubated in 5% CO2 incubator at 37 °C. After 24 h, the attached cancer cells were treated with serial concentrations of CD NPs, ZD NPs, CD, ZD, and standard chemotherapy (fluorouracil) as well as DE, copper chloride and zinc chloride. Following 48 h in 5% CO2 incubator, MTT (5 mg/mL) was incubated with the untreated and treated cells for 4 h then DMSO was added and the absorbance was read at 590 nm43 using spectrophotometer plate reader (BMG LabTech, Germany). The IC50 value, at which 50% cancer growth inhibition, was estimated by GraphPad Prism 9. Additionally, a change in the morphology of the treated breast cancer cell was observed using phase contrast inverted microscope with a digital camera (Olympus, Japan).
Flow cytometry analysis of apoptosis with morphological detection using fluorescence microscopy
After 48 h treatment of MDA-MB 231 with CD NPs, ZD NPs, CD, ZD, and fluorouracil (Fu) at the lowest IC50 (30 μg/mL), these cells were trypsinized, washed, and incubated for 15 min with fluorescein isothiocyanate (FITC)-annexin V/propidium iodide (PI). The percentage of annexin V-stained apoptotic cells was assessed by FITC signal sensor (FL1) versus the phycoerythrin emission signal sensor (FL2)44. Moreover, fluorescence microscope was used after staining adherent MDA-MB 231 cells with acridine orange and ethidium bromide (AO/EB) to confirm the induction of apoptosis in the treated cells.
Investigation of anti-migration efficacy by wound-healing assay
After MDA-MB 231 had reached 90% confluence, the monolayer was scratched and treated with safe doses of the prepared nanocomplexes or traditional complexes (at ~ 0.1 μg/mL). At 0 h and 24 h, the wound area was photographed and assessed by image j software to calculate the percentage inhibition of migration in the treated versus untreated wells.
Determination of ALDH1 activity inhibition
The inhibitory potency (%) of the tested complexes on ALDH1 activity was determined by first incubating (10 min) the untreated and treated cell lysates with 500 µM NAD+ (ALDH cofactor) at 37 °C. Then to initiate ALDH reaction, retinal (substrate) was added. The ALDH activity was assessed as the rate of NADH generation/min/total protein level at 340 nm45 using microplate spectrophotometer (BMG LabTech, Germany). Cellular protein content was quantified using Bradford assay46.
In vivo investigation of anti-metastatic breast tumor potential
Experimental design
The current animal study was conducted upon the agreement of Institutional Animal Care and Use Committee (Alex-IACUC)-member of International Council for Laboratory Animal Science (ICLAS) with approval number AU-0122172832. In accordance with ARRIVE guidelines, all applicable national and international rules for the use of experimental animals were followed.
Ehrlich ascites tumor cells were obtained from frozen aliquots, in liquid nitrogen, then subcultured by intraperitoneal inoculations in BALB/c female mice. The cell viability and count were checked using Trypan Blue exclusion method. The inoculum concentration was adjusted to 2 × 106 cells/0.2 mL phosphate-buffered saline (PBS). The metastatic mammary tumor which was induced by a single injection of Ehrlich cells (2 × 106) in thoracic mammary fat pad of 40 BALB/c female mice (16–20 g), was confirmed by histological examination of liver tissues of 4 mice after 2 weeks (Fig. 3a). Thirty-six mice were blindly divided into 4 groups (9 mice/group), including untreated mammary tumor-bearing group (MT), CD NPs (2 mg/kg)-treated MT group (MT-CD NPs), ZD NPs (2 mg/kg)-treated MT group (MT-ZD NPs), and fluorouracil (10 mg/kg)-treated MT group (MT-Fu). Three treated MT groups were injected intravenously (i.v.) three times a week for two weeks. Mammary tumor volume ([tumor width2 × tumor length]/2) was measured, 3 times/week, using a caliper as well as body weight was recorded 3 times/week.
In parallel, 36 healthy normal mice (16–19 g) were also blindly divided into 4 groups (9 mice/group), including normal healthy (N) untreated group, CD NPs (2 mg/kg)-treated group (N-CD NPs), ZD NPs (2 mg/kg)-treated group (N-ZD NPs), and Fu (10 mg/kg)-treated group (N-Fu).
At the end of the experiment (two mice of MT group died), animals were anaesthetized until their respiration ceased by placing them in a tightly closed chamber containing 5% isoflurane. Then the excised tumors and other tissues (liver, lung, brain, and kidney) were collected. Mammary tumors were weighted for estimating the percentage reduction in tumor growth of the treated groups relative to MT group as well as liver weight was measured. Also, blood was collected in heparin and EDTA tubes for assessing the biochemical parameters and complete blood count (CBC), respectively. Part of the tissues was fixed in 10% formalin for histological and immunohistochemical analyses, while the remainder was stored at – 80 °C for biochemical and molecular analyses.
Histological examinations
The fixed tissues were prepared according to the standard histological protocol. Briefly, these tissues were dehydrated, embedded in paraffin and cut into micrometer sections. Regarding histological analysis, these sections were transferred into traditional slides for staining with hematoxylin and eosin (H&E)47.
Atomic absorption spectrometric quantification of biodistribution of CD NPs and ZD NPs
In a separate animal study, ten MT mice were divided, after 4 weeks of orthotopic injection with Ehrlich cells, into two groups (5 mice/group) which were i.v. injected with 0.1 mg CD NPs and 0.1 mg ZD NPs. After 1 and 4 h, blood was collected from the orbital sinus of isoflurane-anesthetized mice. The next day, animals were sacrificed then blood, thoracic mammary tumor, normal inguinal mammary glands, liver, lung, brain, heart, and kidney were collected. Then copper and zinc which correspond to CD NPs and ZD NPs, respectively, were then quantified, in the digested tissues, using graphite atomic absorption (Analytik Jena AG, Germany). After normalization with the detected concentrations of copper and zinc in tissues of 5 untreated MT mice (as blank), the tissue uptake % was assessed relative to the original injected amounts.
Quantification of tissue ROS and reduced glutathione (GSH) levels
The level of ROS was assessed by incubating DCFDA (10 μM) with 100 μL homogenate (0.1 mg in 1 mL of 40 mM Tris–HCl buffer pH 7.4) of mammary and liver tissues of the untreated (MT and N) and all treated groups. After 40 min, the fluorescence intensity of DCF was measured with fluorometer plate reader (BMG LabTech, Germany) at excitation 485 nm and emission 525 nm48.
The GSH level was determined after incubation of tissue homogenates with Ellman's reagent (5,5′-dithiobis2-nitrobenzoic acid). After 10 min, the absorbance of the generated yellow-colored product was measured with spectrophotometric plate reader (BMG LabTech, Germany) at 412 nm49. The total protein level was determined by Bradford assay46.
Immunohistochemical analyses
The fixed tissues were prepared based on the typical immunohistochemistry protocol. Mammary tumor and liver tissue sections were transferred into positively charged slides that had been dried, dewaxed, boiled in citrate buffer (10 mM, pH 6). Following cooling, these slides were incubated separately with primary antibodies of ki-67, CD 44, and caspase 3. After washing and incubating with horseradish peroxidase-conjugated secondary antibody then washing and adding substrate (3,3′ diaminobenzidine), brown-colored product was formed50, recorded by CellSens imaging analysis software of phase-contrast microscope, and analyzed using image j software.
Assessment of anti-metastatic activity via quantification of liver tumor marker
The common liver tumor marker (α-fetoprotein, AFP) was measured in blood samples of the untreated and treated MT groups using the electrochemiluminescence detection kit method.
Relative change in the expression of tumor suppressor gene and oncogenes
The untreated and treated MT groups’ mammary tumor and liver tissues were homogenized in lysis buffer (supplied in kit) with β-mercaptoethanol then total RNA was extracted according to the standard protocol described in the kit. Following RNA purification and quantification, one-step qPCR SYBR green master mix kit was used with the specific primers (Supplementary Table 1) to assess the relative change in the gene expression of p53, BCl2, HIF-α, β-Catenin, Notch1, ATP Binding Cassette Subfamily G Member 2 (ABCG2), VEGF, and MMP9 using equation of 2−ΔΔCt.
Liver and kidney function parameters and CBC
Parameters’ liver function (ALT, AST and albumin) and kidney function (urea and creatinine) were detected in plasma using colorimetric kits as well as complete blood count (CBC) was determined using a hematology analyzer (Mindray, China).
Statistical analysis
Data are demonstrated as mean ± standard error of mean (SEM) and were analyzed by GraphPad Prism 9.3.1. using one-way analysis of variance (ANOVA)-multiple comparison (Dunnett test) and significance was considered at p < 0.05*, < 0.01**, and < 0.001***.
Supplementary Information
Author contributions
M.M.A. established the current study’s concept and experimental design, carried out the majority of the experiments, analysed the data, and wrote the manuscript. E.Z.A.A is in charge of developing the experimental design for inducing metastatic mammary tumor in mice. Both authors revised and approved the final version of the manuscript.
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Data availability
The original data and materials used in the study are available upon reasonable request from the corresponding authors.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-30553-8.
References
- 1.Wang L, Zhang S, Wang X. The metabolic mechanisms of breast cancer metastasis. Front. Oncol. 2021;10:602416. doi: 10.3389/fonc.2020.602416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rashid NS, Grible JM, Clevenger CV, Harrell JC. Breast cancer liver metastasis: Current and future treatment approaches. Clin. Exp. Metast. 2021;38:263–277. doi: 10.1007/s10585-021-10080-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ji L, Fan L, Zhu X, Gao Y, Wang Z. Prognostic score for de novo metastatic breast cancer with liver metastasis and its predictive value of locoregional treatment benefit. Front. Oncol. 2021;11:651636. doi: 10.3389/fonc.2021.651636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tasdogan A, Ubellacker JM, Morrison SJ. Redox regulation in cancer cells during metastasis. Cancer Discov. 2021;11:2682–2692. doi: 10.1158/2159-8290.CD-21-0558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma R, et al. Mechanisms involved in breast cancer liver metastasis. J. Transl. Med. 2015;13:64. doi: 10.1186/s12967-015-0425-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou HM, Zhang JG, Zhang X, Li Q. Targeting cancer stem cells for reversing therapy resistance: Mechanism, signaling, and prospective agents. Sig. Transduct. Target. Ther. 2021;6:62. doi: 10.1038/s41392-020-00430-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kamble D, Mahajan M, Dhat R, Sitasawad S. Keap1-Nrf2 pathway regulates ALDH and contributes to radioresistance in breast cancer stem cells. Cells. 2021;10:83. doi: 10.3390/cells10010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wei Y, et al. ALDH1: A potential therapeutic target for cancer stem cells in solid tumors. Front. Oncol. 2022;12:1026278. doi: 10.3389/fonc.2022.1026278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Croker AK, et al. Differential functional roles of ALDH1A1 and ALDH1A3 in mediating metastatic behavior and therapy resistance of human breast cancer cells. Int. J. Mol. Sci. 2017;18(10):2039. doi: 10.3390/ijms18102039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao T, et al. The mechanism between epithelial mesenchymal transition in breast cancer and hypoxia microenvironment. Biomed. Pharmacother. 2016;80:393–405. doi: 10.1016/j.biopha.2016.02.044. [DOI] [PubMed] [Google Scholar]
- 11.Lu H, et al. Chemotherapy triggers HIF-1-dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc. Natl. Acad. Sci. U.S.A. 2015;112:E4600–E4609. doi: 10.1073/pnas.1513433112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, et al. Twist1 promotes breast cancer invasion and metastasis by silencing Foxa1 expression. Oncogene. 2017;36:1157–1166. doi: 10.1038/onc.2016.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alshaer W, et al. Downregulation of STAT3, β-catenin, and Notch-1 by single and combinations of siRNA treatment enhance chemosensitivity of wild type and doxorubicin resistant MCF7 breast cancer cells to doxorubicin. Int. J. Mol. Sci. 2019;20:3696. doi: 10.3390/ijms20153696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takebe N, et al. Targeting Notch, Hedgehog, and Wnt pathways in cancer stem cells: Clinical update. Nat. Rev. Clin. Oncol. 2015;12:445–464. doi: 10.1038/nrclinonc.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao J, Jin Q, Wang XD, Zhu HJ, Ni QC. Aldehyde dehydrogenase 1 expression is correlated with poor prognosis in breast cancer. Medicine (Baltimore) 2017;96(25):e7171. doi: 10.1097/MD.0000000000007171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srikoon P, et al. Diethyldithiocarbamate suppresses an NF-kappaB dependent metastatic pathway in cholangiocarcinoma cells. Asian Pac. J. Cancer Prev. 2013;14:4441–4446. doi: 10.7314/APJCP.2013.14.7.4441. [DOI] [PubMed] [Google Scholar]
- 17.Bu W, et al. Disulfiram inhibits epithelial-mesenchymal transition through TGFβ-ERK-snail pathway independently of Smad4 to decrease oral squamous cell carcinoma metastasis. Cancer Manag. Res. 2019;11:3887–3898. doi: 10.2147/CMAR.S199912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren L, et al. Diethyldithiocarbamate-copper nanocomplex reinforces disulfiram chemotherapeutic efficacy through light-triggered nuclear targeting. Theranostics. 2020;10:6384–6398. doi: 10.7150/thno.45558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skrott Z, et al. Alcohol-abuse drug disulfiram targets cancer via p97 segregase adaptor NPL4. Nature. 2017;552:194–199. doi: 10.1038/nature25016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cvek B, Milacic V, Taraba J, Dou QP. Ni(II), Cu(II), and Zn(II) diethyldithiocarbamate complexes show various activities against the proteasome in breast cancer cells. J. Med. Chem. 2008;51:6256–6258. doi: 10.1021/jm8007807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahmood RI, et al. Biosynthesis of copper oxide nanoparticles mediated Annona muricata as cytotoxic and apoptosis inducer factor in breast cancer cell lines. Sci. Rep. 2022;12(1):16165. doi: 10.1038/s41598-022-20360-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mohammed SAA, et al. Copper oxide nanoparticle-decorated carbon nanoparticle composite colloidal preparation through laser ablation for antimicrobial and antiproliferative actions against breast cancer cell line, MCF-7. Biomed. Res. Int. 2022;2022:9863616. doi: 10.1155/2022/9863616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.George BP, Rajendran NK, Houreld NN, Abrahamse H. Rubus capped zinc oxide nanoparticles induce apoptosis in MCF-7 breast cancer cells. Molecules. 2022;27(20):6862. doi: 10.3390/molecules27206862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loganathan S, Shivakumar MS, Karthi S, Nathan SS, Selvam K. Metal oxide nanoparticle synthesis (ZnO-NPs) of Knoxia sumatrensis (Retz.) DC. Aqueous leaf extract and it’s evaluation of their antioxidant, anti-proliferative and larvicidal activities. Toxicol. Rep. 2021;8:64–72. doi: 10.1016/j.toxrep.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shamasi Z, Es-haghi A, Taghavizadeh Yazdi ME, Amiri MS, Homayouni-Tabrizi M. Role of Rubia tinctorum in the synthesis of zinc oxide nanoparticles and apoptosis induction in breast cancer cell line. Nanomed. J. 2021;8:65–72. [Google Scholar]
- 26.Karlsson HL, Cronholm P, Gustafsson J, Moller L. Copper oxide nanoparticles are highly toxic: A comparison between metal oxide nanoparticles and carbon nanotubes. Chem. Res. Toxicol. 2008;21:1726–1732. doi: 10.1021/tx800064j. [DOI] [PubMed] [Google Scholar]
- 27.Abu-Serie MM, Eltarahony M. Novel nanoformulated diethyldithiocarbamate complexes with biosynthesized or green chemosynthesized copper oxide nanoparticles: An in vitro comparative anticancer study. Int. J. Pharm. 2021;609:121149. doi: 10.1016/j.ijpharm.2021.121149. [DOI] [PubMed] [Google Scholar]
- 28.Koppaka V, et al. Aldehyde dehydrogenase inhibitors: A comprehensive review of the pharmacology, mechanism of action, substrate specificity, and clinical application. Pharmacol. Rev. 2012;64:520–539. doi: 10.1124/pr.111.005538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skrott Z, Cvek B. Diethyldithiocarbamate complex with copper: The mechanism of action in cancer cells. Mini Rev. Med. Chem. 2012;12:1184–1192. doi: 10.2174/138955712802762068. [DOI] [PubMed] [Google Scholar]
- 30.Thanuja J, Udayabhanu A, Nagaraju G, Naika HR. Biosynthesis of Cu4O3 nanoparticles using Razma seeds: Application to antibacterial and cytotoxicity activities. SN Appl. Sci. 2019;1:1646. doi: 10.1007/s42452-019-1556-3. [DOI] [Google Scholar]
- 31.Husain N, Mahmood R. Copper(II) generates ROS and RNS, impairs antioxidant system and damages membrane and DNA in human blood cells. Environ. Sci. Pollut. Res. Int. 2019;26:20654–20668. doi: 10.1007/s11356-019-05345-1. [DOI] [PubMed] [Google Scholar]
- 32.Coyle P, Philcox JC, Carey LC, Rofe AM. Metallothionein: The multipurpose protein. Cell Mol. Life Sci. 2002;59:627–647. doi: 10.1007/s00018-002-8454-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Soni D, Gandhi D, Tarale P, Bafana A, Pandey RA, Sivanesan S. Oxidative stress and genotoxicity of zinc oxide nanoparticles to pseudomonas species, human promyelocytic leukemic (HL-60), and blood cells. Biol. Trace Elem. Res. 2017;178:218–227. doi: 10.1007/s12011-016-0921-y. [DOI] [PubMed] [Google Scholar]
- 34.Kalyane D, Raval N, Maheshwari R, Tambe V, Kalia K, Tekade RK. Employment of enhanced permeability and retention effect (EPR): Nanoparticle-based precision tools for targeting of therapeutic and diagnostic agent in cancer. Mater. Sci. Eng. C Mater. Biol. Appl. 2019;98:1252–1276. doi: 10.1016/j.msec.2019.01.066. [DOI] [PubMed] [Google Scholar]
- 35.Zein R, Sharrouf W, Selting K. Physical properties of nanoparticles that result in improved cancer targeting. J. Oncol. 2020;2020:5194780. doi: 10.1155/2020/5194780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Redza-Dutordoir M, Averill-Bates DA. Activation of apoptosis signalling pathways by reactive oxygen species. Biochim. Biophys. Acta. 2016;1863:2977–2992. doi: 10.1016/j.bbamcr.2016.09.012. [DOI] [PubMed] [Google Scholar]
- 37.Luo M, Wicha MS. Targeting cancer stem cell redox metabolism to enhance therapy responses. Semin. Radiat. Oncol. 2019;29:42–54. doi: 10.1016/j.semradonc.2018.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu J, Wang Z. Increased oxidative stress as a selective anticancer therapy. Oxid. Med. Cell Longev. 2015;2015:294303. doi: 10.1155/2015/294303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng Q, Zhang M, Zhou F, Zhang L, Meng X. The breast cancer stem cells traits and drug resistance. Front. Pharmacol. 2021;11:599965. doi: 10.3389/fphar.2020.599965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen H, et al. Inhibiting the PI3K/AKT/mTOR signalling pathway with copper oxide nanoparticles from Houttuynia cordata plant: Attenuating the proliferation of cervical cancer cells. Artif. Cells Nanomed. Biotechnol. 2021;49:240–249. doi: 10.1080/21691401.2021.1890101. [DOI] [PubMed] [Google Scholar]
- 41.Yang Y, Deng Q, Feng X, Sun J. Use of the disulfiram/copper complex for breast cancer chemoprevention in MMTV-erbB2 transgenic mice. Mol. Med. Rep. 2015;12:746–752. doi: 10.3892/mmr.2015.3426. [DOI] [PubMed] [Google Scholar]
- 42.Chaithanatkun, N., Chantarawong, D., Songkeaw, P., Onlaor, K., Thiwawong, T. & Tunhoo, B. Effect of ascorbic acid on structural properties of ZnO nanoparticles prepared by precipitation process. In 10th IEEE International Conference on Nano/Micro Engineered and Molecular Systems 145–148 (2015).
- 43.Mosmann T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 44.Al-Salman HNK, Ali ET, Jabir M, Sulaiman GM, Al-Jadaan SA. 2-Benzhydrylsulfinyl-N-hydroxyacetamide-Na extracted from fig as a novel cytotoxic and apoptosis inducer in SKOV-3 and AMJ-13 cell lines via P53 and caspase-8 pathway. Eur. Food Res. Technol. 2020;246(8):1591–1608. doi: 10.1007/s00217-020-03515-x. [DOI] [Google Scholar]
- 45.Graham CE, Brocklehurst K, Pickersgill RW, Warren MJ. Characterization of retinaldehyde dehydrogenase 3. Biochem. J. 2006;394:67–75. doi: 10.1042/BJ20050918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 47.Cardiff RD, Miller CH, Munn RJ. Manual hematoxylin and eosin staining of mouse tissue sections. Cold Spring Harb. Protoc. 2014;2014(6):655–658. doi: 10.1101/pdb.prot073411. [DOI] [PubMed] [Google Scholar]
- 48.Caro AA, Adlong LW, Crocker SJ, Gardner MW, Luikart EF, Gron LU. Effect of garlic derived organosulfur compounds on mitochondrial function and integrity in isolated mouse liver mitochondria. Toxicol. Lett. 2012;214:166–174. doi: 10.1016/j.toxlet.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ellman GL. Tissue sulfhydryl groups. Arch. Biochem. Biophys. 1959;82:70–77. doi: 10.1016/0003-9861(59)90090-6. [DOI] [PubMed] [Google Scholar]
- 50.Shi SR, Liu C, Taylor CR. Standardization of immunohistochemistry for formalin-fixed, paraffin-embedded tissue sections based on the antigen-retrieval technique: From experiments to hypothesis. J. Histochem. Cytochem. 2007;55:105–109. doi: 10.1369/jhc.6P7080.2006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original data and materials used in the study are available upon reasonable request from the corresponding authors.