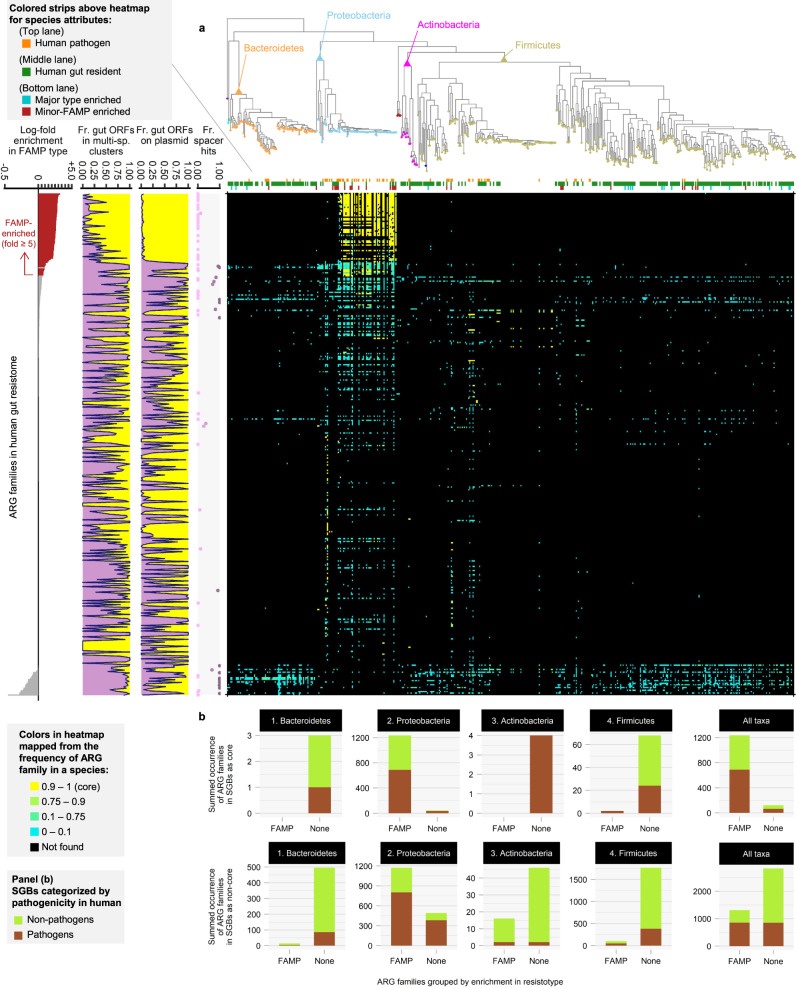
Fig. 4. Phylogenetic distribution of the ARG families in the two resistotypes.
Main panel (a): heat map gives presence of each ARG family across the phylogenetic tree of the SGBs, color reflects the fraction of genomes in the SGB that contain the ARG family. The 363 ARG families detected in adult stool metagenomes based on our assembled catalogue are sorted (y-axis) by fold-difference between the mean abundances in the background and FAMP resistotypes. The phylogenetic tree includes the 522 SGBs which are most abundant in the gut microbiome or are associated with resistotypes (see Methods). Subpanels left to right: bar plot of the fold-difference in abundance between the resistotypes; fraction of ORFs in multi-species ARG_cluster99s in each ARG family; fraction of plasmid-borne ORFs; fraction of ORFs containing at least one high-identity alignment of CRISPR spacers collected from gut metagenomes (minimum 90% identity over 90% of the spacer length) in each ARG family. Panel (b): For each major phylum and for all taxa combined, we give the total occurrence of ARG families in the core resistomes of the SGBs (top row) and the accessory resistomes (bottom row). FAMP-associated and non-associated ARG families were compared, and the occurrence in pathogenic and non-pathogenic species were coloured differently. The maximum-likelihood phylogenetic tree was reconstructed using one representative genome for each of the selected SGBs. Concatenated nucleotide sequence alignments of 40 single-copy COGs were used as the input. The overall contribution of ARG subsets based on pathogenicity and residence to the resistomes of background and FAMP reistotypes is summarized in Fig. S10.