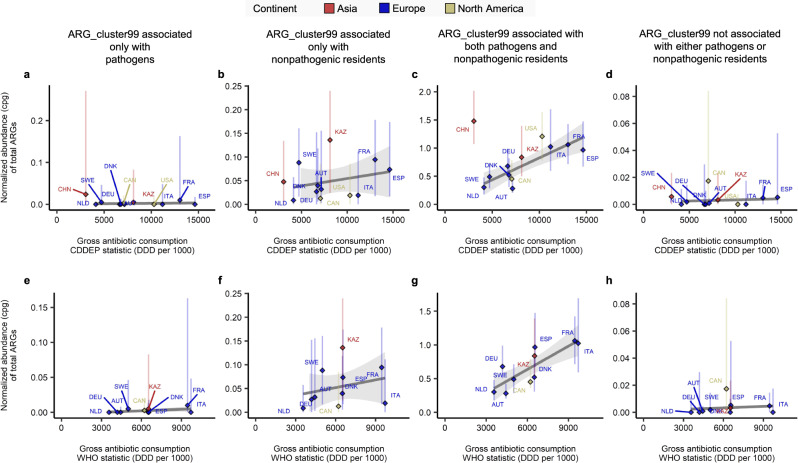
Fig. 5. Correlation between median total abundance of ARGs deriving from different species types and antibiotic consumption rates.
ARG_cluster99s were assigned to four categories based on the presence of ORFs from pathogens (see definition in text) and non-pathogenic residents (non-pathogenic found in at least 10% of the stool metagenomes of healthy adults not taking antibiotics). a, b, c, d We display correlations between the median ARG abundance (cpg) summed for each species category and the per capita rate of antibiotic consumption (DDD per 1000) in each country using CDDEP statistics. Pearson’s correlation a r = 0.19, p = 0.58; b r = 0.25, p = 0.46; c r = 0.81, p = 2.6e-3; d r = 0.11, p = 0.76. In e, f, g, h we used the WHO antibiotic consumption. Pearson’s correlation e r = 0.44 p = 0.2; f r = 0.26, p = 0.61; g r = 0.81, p = 4.2e-3; h r = 0.14, p = 0.69. Diamonds give the medians of the countries. Vertical lines indicate the range from the first quartile to the third quartile. Linear trend line was determined by generalized linear model using ggplot2 R package, shaded area represents 95% confidence interval. The number of metagenome samples used to derive the median and the range bar shown for each country in A-H: AUT (n = 16), CAN (n = 36), CHN (n = 340), DEU (n = 103), DNK (n = 401), ESP (n = 139), FRA (n = 62), ITA (n = 33), KAZ (n = 168), NLD (n = 470), SWE (n = 109), USA (n = 147). Correlation test statistics are given in Table S4. The numbers summarizing two-way categorization of species are provided in Table S9. Comparisons between the results from detection schemes of plasmid-borne ARGs and multi-species ARGs, and between the results from analyzing gut metagenomic ORFs and RefSeq genomic ORFs are provided in Fig. S10.