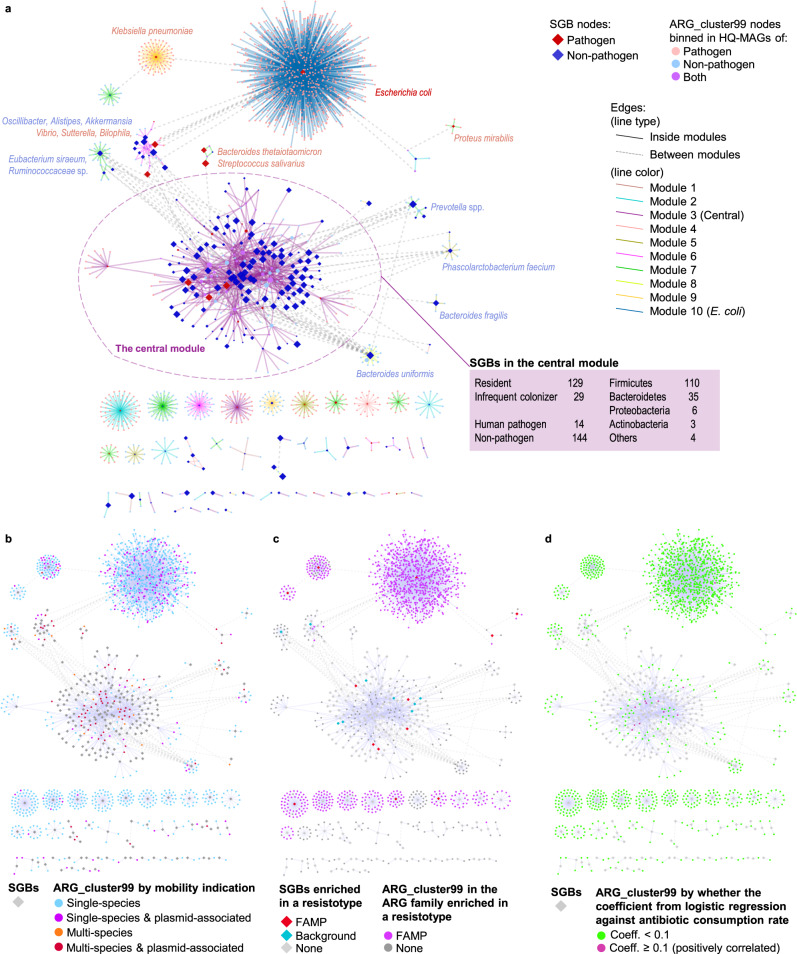
Fig. 7. Bipartite network of ARG clusters and species-level genomic bins based on the genomes reconstructed from stool metagenomes.
Network comprising species-level genomic bins (SGBs) connected to ARG_cluster99s they contain (ORF frequency > 1) generated from high-quality MAGs (HQ-MAGs) constructed from 6104 adult stool metagenomes. Ten modules in the network were detected using Beckett’s method (computeModules of the R package bipartite). The largest (SGBs n = 158) was defined as `central' the others all (n < 20) as peripheral. a SGB and ARG_cluster99 nodes coloured by pathogen status, edges by module (when shared). b SGB not coloured, ARG_cluster99s by single-species, multi-species, plasmid-borne. c SGB nodes and ARG_cluster99s were categorised into background- or FAMP-associated. d SGB nodes were not categorised, ARG_cluster99s categorised by association with country-level antibiotic consumption (see Methods).